7. Tabular muris (FACS - 10x) integration analysis#
import os
import glob
import sys
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.manifold import TSNE
from umap import UMAP
import seaborn as sns
from itertools import compress
#sys.path.append('.')
import src.utils as my_u
from src.utils import df_cp
from src.utils import df_log
from src.utils import df_total20000
from src.utils import df_minmax
from src.utils import df_minmax_scaler
from src.utils import df_l2norm
from src.utils import df_zscore
from src.utils import df_meansquare
from src.utils import run_plot
import random
import scanpy
2023-10-05 14:24:49.342602: I tensorflow/tsl/cuda/cudart_stub.cc:28] Could not find cuda drivers on your machine, GPU will not be used.
2023-10-05 14:24:49.854306: I tensorflow/tsl/cuda/cudart_stub.cc:28] Could not find cuda drivers on your machine, GPU will not be used.
2023-10-05 14:24:49.856223: I tensorflow/core/platform/cpu_feature_guard.cc:182] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.
To enable the following instructions: AVX2 FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.
2023-10-05 14:24:51.690830: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT
Common organ in Tabula Muris FACS & 10x#
tabula_file_list = glob.glob("./dataset/00_facs_raw_data/FACS/*.csv")
print(len(tabula_file_list))
#print(tabula_file_list)
# Tabula data load
#
tabula_labels = []
for ff in tabula_file_list:
tabula_labels += [ff.split('/')[-1].split('-')[0]]
tabula_labels_set = set(tabula_labels)
print(tabula_labels_set)
20
{'Large_Intestine', 'Mammary_Gland', 'Diaphragm', 'Marrow', 'Aorta', 'Brain_Non', 'Thymus', 'Skin', 'Pancreas', 'Bladder', 'Trachea', 'Kidney', 'Liver', 'Brain_Myeloid', 'Tongue', 'Limb_Muscle', 'Lung', 'Heart', 'Fat', 'Spleen'}
tabula_10xfile_list = glob.glob("./dataset/01_droplet_raw_data/droplet/*")
print(len(tabula_10xfile_list))
#print(tabula_10xfile_list)
# Tabula data load
tabula_10x_labels = []
for ff in tabula_10xfile_list:
tabula_10x_labels += [ff.split('/')[-1].split('-')[0]]
tabula_10x_labels_set = set(tabula_10x_labels)
print(tabula_10x_labels_set)
28
{'Mammary_Gland', 'Limb_Muscle', 'Heart_and_Aorta', 'Lung', 'Marrow', 'Trachea', 'Thymus', 'Tongue', 'Kidney', 'Spleen', 'Bladder', 'Liver'}
set(tabula_labels_set) & set(tabula_10x_labels_set)
{'Bladder',
'Kidney',
'Limb_Muscle',
'Liver',
'Lung',
'Mammary_Gland',
'Marrow',
'Spleen',
'Thymus',
'Tongue',
'Trachea'}
mouse_label = pd.read_csv('./dataset/01_droplet_raw_data/metadata_droplet.csv', sep=',', header=0)
mouse_dic = dict(mouse_label[['channel','mouse.id']].values)
mouse_dic
{'10X_P4_0': '3-M-8',
'10X_P4_1': '3-M-9',
'10X_P4_2': '3-M-8/9',
'10X_P4_3': '3-M-8',
'10X_P4_4': '3-M-9',
'10X_P4_5': '3-M-8',
'10X_P4_6': '3-M-9',
'10X_P4_7': '3-M-8',
'10X_P7_0': '3-F-56',
'10X_P7_1': '3-F-57',
'10X_P7_2': '3-F-56',
'10X_P7_3': '3-F-57',
'10X_P7_4': '3-F-56',
'10X_P7_5': '3-F-57',
'10X_P7_6': '3-F-56',
'10X_P7_7': '3-F-56',
'10X_P7_8': '3-F-56',
'10X_P7_9': '3-F-57',
'10X_P7_10': '3-F-56',
'10X_P7_11': '3-F-56',
'10X_P7_12': '3-F-56',
'10X_P7_13': '3-F-57',
'10X_P7_14': '3-F-56',
'10X_P7_15': '3-F-57',
'10X_P8_12': '3-M-5/6',
'10X_P8_13': '3-M-7/8',
'10X_P8_14': '3-M-5/6',
'10X_P8_15': '3-M-7/8'}
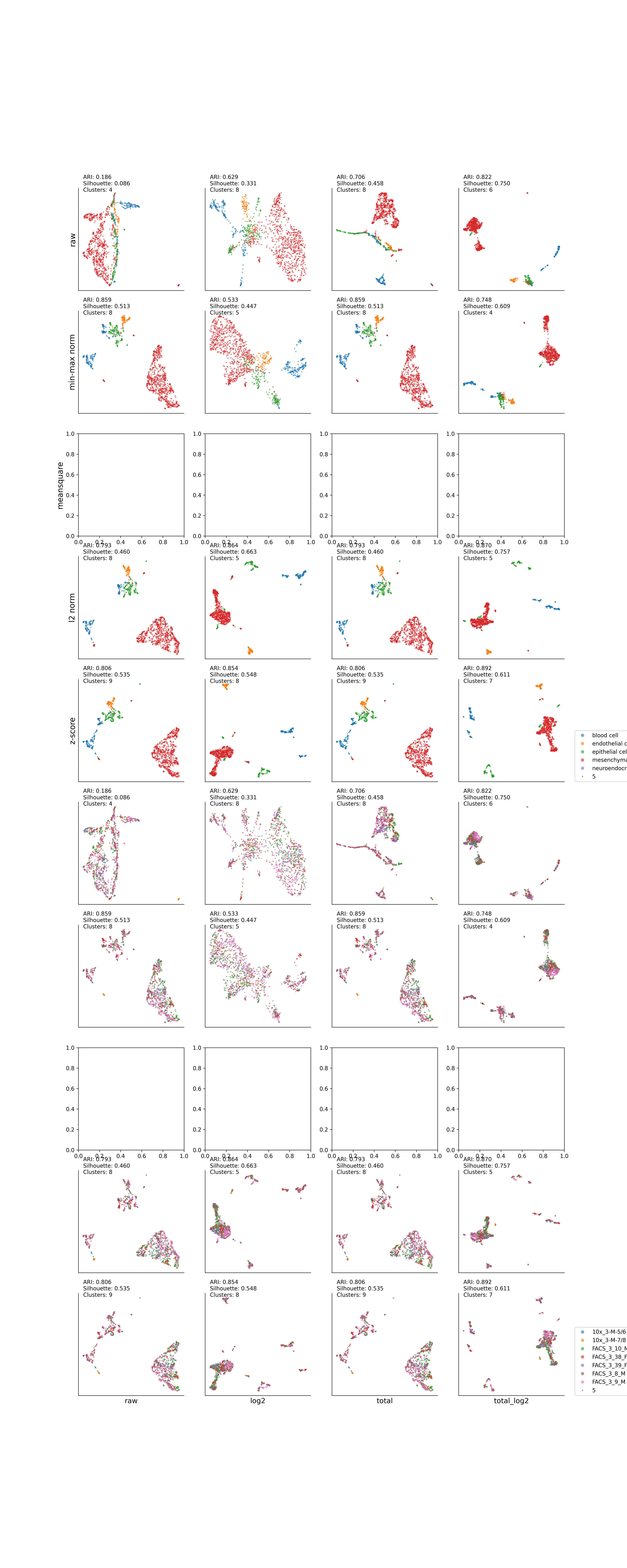
Analyzed organs {‘Bladder’, ‘Kidney’, ‘Limb_Muscle’, ‘Liver’, ‘Lung’, ‘Mammary_Gland’, ‘Marrow’, ‘Spleen’, ‘Thymus’, ‘Tongue’, ‘Trachea’}
Bladder UMAP:DBSCAN results#
ORGAN = 'Bladder'
tabula_file_list = glob.glob("../data/tabular_muris/00_facs_raw_data/FACS/*"+ORGAN+"*.csv")
print(tabula_file_list)
tabula_10xfile_list = glob.glob("../data/tabular_muris/01_droplet_raw_data/droplet/*"+ORGAN+"*")
print(tabula_10xfile_list)
# Tabula data load
#
tabula_labels = []
tabula_data = pd.DataFrame()
for ff in tabula_file_list:
data = pd.read_csv(ff, sep=',', index_col=0, header=0)
data = data.transpose()
c = list(range(data.shape[0]))
tabula_data = pd.concat([tabula_data, data.iloc[c,]], axis=0)
tabula_labels += [ff.rstrip('-counts.csv').split('/')[-1]] * data.shape[0]
print(tabula_data.shape)
tabula_labels_set = set(tabula_labels)
# Tabula label load
#
annot_label = pd.read_csv('../data/tabular_muris/00_facs_raw_data/annotations_FACS.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_data.index)
m = annot_label.filter(com, axis=0)
tabula_data = tabula_data.filter(com, axis=0)
tabula_data = pd.concat([tabula_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue = tabula_data['tissue']
tabula_labels = tabula_data['cell_ontology_class']
tissue = tissue.values.tolist()
tabula_labels = tabula_labels.values.tolist()
tabula_data.pop('cell_ontology_class')
tabula_data.pop('tissue')
tabula_data.pop('zsGreen_transgene')
print(set(tabula_labels))
# Tabula 10x data load
#
tabula_10x_batches = []
tabula_10x_data = pd.DataFrame()
for ff in tabula_10xfile_list:
data = scanpy.read_10x_mtx(ff)
data = data.to_df()
prefix_cell = ff.split('-')[1]
data.index = [prefix_cell +'_'+x.split('-')[0] for x in data.index]
tabula_10x_data = pd.concat([tabula_10x_data, data], axis=0)
tabula_10x_batches += [ff.split('/')[-1].split('-')[1]] * data.shape[0]
print(tabula_10x_data.shape)
# Tabula 10x label load
#
annot_label = pd.read_csv('../data/tabular_muris/01_droplet_raw_data/annotations_droplet.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_10x_data.index)
m = annot_label.filter(com, axis=0)
tabula_10x_data = tabula_10x_data.filter(com, axis=0)
tabula_10x_data = pd.concat([tabula_10x_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue_10x = tabula_10x_data['tissue']
tabula_10x_labels = tabula_10x_data['cell_ontology_class']
tissue_10x = tissue_10x.values.tolist()
tabula_10x_labels = tabula_10x_labels.values.tolist()
tabula_10x_data.pop('cell_ontology_class')
tabula_10x_data.pop('tissue')
tabula_10x_data.pop('zsGreen_transgene')
print(set(tabula_10x_labels))
total_data = pd.concat([tabula_data, tabula_10x_data], axis=0)
labels = tissue + tissue_10x #mca_labels
labels_cell = tabula_labels + tabula_10x_labels #mca_labels
blabels = ['FACS_'+x.split('.')[2] for x in tabula_data.index] + ['10x_'+mouse_dic['_'.join(x.split('_')[0:3])] for x in tabula_10x_data.index]
total_data = total_data.replace(np.NaN,0)
print('\nSummary')
print(total_data.shape)
print(set(labels))
print(set(labels_cell))
print(set(blabels))
#latent_space = TSNE(n_components=2)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
clustering_method = 'dbscan'
############################################
plt.figure(figsize=(16,40), dpi=300)
ax00 = plt.subplot2grid((10,4), (0,0))
ax10 = plt.subplot2grid((10,4), (0,1))
ax20 = plt.subplot2grid((10,4), (0,2))
ax30 = plt.subplot2grid((10,4), (0,3))
ax01 = plt.subplot2grid((10,4), (1,0))
ax11 = plt.subplot2grid((10,4), (1,1))
ax21 = plt.subplot2grid((10,4), (1,2))
ax31 = plt.subplot2grid((10,4), (1,3))
ax02 = plt.subplot2grid((10,4), (2,0))
ax12 = plt.subplot2grid((10,4), (2,1))
ax22 = plt.subplot2grid((10,4), (2,2))
ax32 = plt.subplot2grid((10,4), (2,3))
ax03 = plt.subplot2grid((10,4), (3,0))
ax13 = plt.subplot2grid((10,4), (3,1))
ax23 = plt.subplot2grid((10,4), (3,2))
ax33 = plt.subplot2grid((10,4), (3,3))
ax04 = plt.subplot2grid((10,4), (4,0))
ax14 = plt.subplot2grid((10,4), (4,1))
ax24 = plt.subplot2grid((10,4), (4,2))
ax34 = plt.subplot2grid((10,4), (4,3))
ax05 = plt.subplot2grid((10,4), (5,0))
ax15 = plt.subplot2grid((10,4), (5,1))
ax25 = plt.subplot2grid((10,4), (5,2))
ax35 = plt.subplot2grid((10,4), (5,3))
ax06 = plt.subplot2grid((10,4), (6,0))
ax16 = plt.subplot2grid((10,4), (6,1))
ax26 = plt.subplot2grid((10,4), (6,2))
ax36 = plt.subplot2grid((10,4), (6,3))
ax07 = plt.subplot2grid((10,4), (7,0))
ax17 = plt.subplot2grid((10,4), (7,1))
ax27 = plt.subplot2grid((10,4), (7,2))
ax37 = plt.subplot2grid((10,4), (7,3))
ax08 = plt.subplot2grid((10,4), (8,0))
ax18 = plt.subplot2grid((10,4), (8,1))
ax28 = plt.subplot2grid((10,4), (8,2))
ax38 = plt.subplot2grid((10,4), (8,3))
ax09 = plt.subplot2grid((10,4), (9,0))
ax19 = plt.subplot2grid((10,4), (9,1))
ax29 = plt.subplot2grid((10,4), (9,2))
ax39 = plt.subplot2grid((10,4), (9,3))
l = []
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_cp(total_data), \
ax00, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax05)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_cp(total_data)), \
ax10, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax15)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_total20000(df_cp(total_data)), \
ax20, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax25)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_total20000(df_cp(total_data))), \
ax30, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax35)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_cp(total_data)), \
ax01, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax06)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_cp(total_data))), \
ax11, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax16)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_total20000(df_cp(total_data))), \
ax21, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax26)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_total20000(df_cp(total_data)))), \
ax31, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax36)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_cp(total_data)), \
ax02, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax07)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_cp(total_data))), \
ax12, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax17)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_total20000(df_cp(total_data))), \
ax22, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax27)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_total20000(df_cp(total_data)))), \
ax32, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax37)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_cp(total_data)), \
ax03, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax08)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_cp(total_data))), \
ax13, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax18)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_total20000(df_cp(total_data))), \
ax23, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax28)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_total20000(df_cp(total_data)))), \
ax33, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax38)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_cp(total_data)), \
ax04, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax09)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_cp(total_data))), \
ax14, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax19)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_total20000(df_cp(total_data))), \
ax24, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax29)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_total20000(df_cp(total_data)))), \
ax34, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax39)
)
############################################
ax00.set_ylabel('raw' , fontsize=14)
ax01.set_ylabel('min-max norm' , fontsize=14)
ax02.set_ylabel('meansquare' , fontsize=14)
ax03.set_ylabel('l2 norm' , fontsize=14)
ax04.set_ylabel('z-score' , fontsize=14)
ax09.set_xlabel('raw', fontsize=13)
ax19.set_xlabel('log2', fontsize=13)
ax29.set_xlabel('total', fontsize=13)
ax39.set_xlabel('total_log2', fontsize=13)
ax34.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
ax39.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
['../data/tabular_muris/00_facs_raw_data/FACS/Bladder-counts.csv']
['../data/tabular_muris/01_droplet_raw_data/droplet/Bladder-10X_P4_4', '../data/tabular_muris/01_droplet_raw_data/droplet/Bladder-10X_P7_7', '../data/tabular_muris/01_droplet_raw_data/droplet/Bladder-10X_P4_3']
(1638, 23433)
{'basal cell of urothelium', 'mesenchymal cell', 'bladder cell'}
(1783, 23433)
(2351, 23433)
(2500, 23433)
{'basal cell of urothelium', 'endothelial cell', 'mesenchymal cell', 'bladder cell', 'leukocyte'}
Summary
(3787, 23432)
{'Bladder'}
{'basal cell of urothelium', 'endothelial cell', 'mesenchymal cell', 'bladder cell', 'leukocyte'}
{'FACS_3_10_M', 'FACS_3_8_M', 'FACS_3_39_F', '10x_3-F-56', 'FACS_3_56_F', 'FACS_3_9_M', '10x_3-M-9', '10x_3-M-8', 'FACS_3_38_F'}
dbscan #cluster: (5, 0.34725168975256304, 0.38760608, 0.26821518523129695)
dbscan #cluster: (2, 0.7470687973729015, 0.68757266, 0.0035017157527061724)
dbscan #cluster: (6, 0.4778870399984946, 0.72933733, 0.29361421024388196)
dbscan #cluster: (4, 0.706017167418482, 0.7751131, 0.13012743380889571)
dbscan #cluster: (5, 0.459531830238682, 0.7658125, 0.30038063461847664)
dbscan #cluster: (4, 0.8122298200173916, 0.7340923, -0.0019745126682521597)
dbscan #cluster: (5, 0.459531830238682, 0.7658125, 0.30038063461847664)
dbscan #cluster: (5, 0.7193953588947256, 0.6713893, 0.12086698601593499)
dbscan #cluster: (7, 0.4737749344572787, 0.46333757, 0.2986158400188783)
dbscan #cluster: (3, 0.7798019064211744, 0.7031068, -0.0009266209050112834)
dbscan #cluster: (7, 0.4737749344572787, 0.46333757, 0.2986158400188783)
dbscan #cluster: (6, 0.47913904522939965, 0.8315169, 0.29514642238069605)
dbscan #cluster: (7, 0.47471253404888586, 0.65699065, 0.29829815198210197)
dbscan #cluster: (2, 0.7477635833627887, 0.7832111, 0.003457735760287989)
dbscan #cluster: (7, 0.47471253404888586, 0.65699065, 0.29829815198210197)
dbscan #cluster: (6, 0.47913904522939965, 0.8501285, 0.29514642238069605)
dbscan #cluster: (7, 0.4735622528618166, 0.6219761, 0.29890741693328526)
dbscan #cluster: (4, 0.808081027973932, 0.7203113, -0.0013152971626582191)
dbscan #cluster: (7, 0.4735622528618166, 0.6219761, 0.29890741693328526)
dbscan #cluster: (6, 0.47949506016600374, 0.8522924, 0.29483626411420605)
<matplotlib.legend.Legend at 0x7fcfd176bf10>

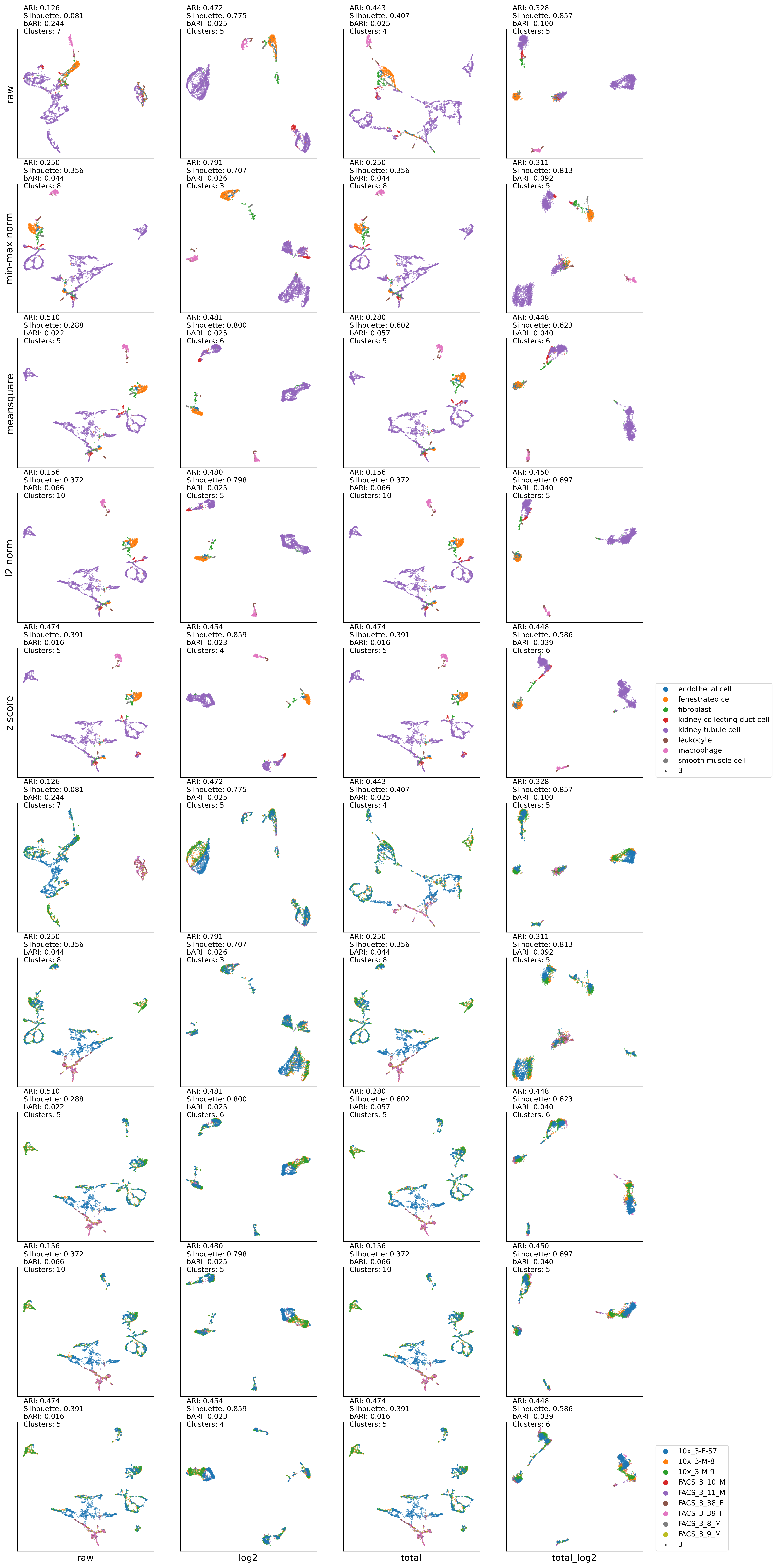
Kidney UMAP:DBSCAN results#
ORGAN = 'Kidney'
tabula_file_list = glob.glob("../data/tabular_muris/00_facs_raw_data/FACS/*"+ORGAN+"*.csv")
print(tabula_file_list)
tabula_10xfile_list = glob.glob("../data/tabular_muris/01_droplet_raw_data/droplet/*"+ORGAN+"*")
print(tabula_10xfile_list)
# Tabula data load
#
tabula_labels = []
tabula_data = pd.DataFrame()
for ff in tabula_file_list:
data = pd.read_csv(ff, sep=',', index_col=0, header=0)
data = data.transpose()
c = list(range(data.shape[0]))
tabula_data = pd.concat([tabula_data, data.iloc[c,]], axis=0)
tabula_labels += [ff.rstrip('-counts.csv').split('/')[-1]] * data.shape[0]
print(tabula_data.shape)
tabula_labels_set = set(tabula_labels)
# Tabula label load
#
annot_label = pd.read_csv('../data/tabular_muris/00_facs_raw_data/annotations_FACS.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_data.index)
m = annot_label.filter(com, axis=0)
tabula_data = tabula_data.filter(com, axis=0)
tabula_data = pd.concat([tabula_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue = tabula_data['tissue']
tabula_labels = tabula_data['cell_ontology_class']
tissue = tissue.values.tolist()
tabula_labels = tabula_labels.values.tolist()
tabula_data.pop('cell_ontology_class')
tabula_data.pop('tissue')
tabula_data.pop('zsGreen_transgene')
print(set(tabula_labels))
# Tabula 10x data load
#
tabula_10x_batches = []
tabula_10x_data = pd.DataFrame()
for ff in tabula_10xfile_list:
data = scanpy.read_10x_mtx(ff)
data = data.to_df()
prefix_cell = ff.split('-')[1]
data.index = [prefix_cell +'_'+x.split('-')[0] for x in data.index]
tabula_10x_data = pd.concat([tabula_10x_data, data], axis=0)
tabula_10x_batches += [ff.split('/')[-1].split('-')[1]] * data.shape[0]
print(tabula_10x_data.shape)
# Tabula 10x label load
#
annot_label = pd.read_csv('../data/tabular_muris/01_droplet_raw_data/annotations_droplet.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_10x_data.index)
m = annot_label.filter(com, axis=0)
tabula_10x_data = tabula_10x_data.filter(com, axis=0)
tabula_10x_data = pd.concat([tabula_10x_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue_10x = tabula_10x_data['tissue']
tabula_10x_labels = tabula_10x_data['cell_ontology_class']
tissue_10x = tissue_10x.values.tolist()
tabula_10x_labels = tabula_10x_labels.values.tolist()
tabula_10x_data.pop('cell_ontology_class')
tabula_10x_data.pop('tissue')
tabula_10x_data.pop('zsGreen_transgene')
print(set(tabula_10x_labels))
total_data = pd.concat([tabula_data, tabula_10x_data], axis=0)
labels = tissue + tissue_10x #mca_labels
labels_cell = tabula_labels + tabula_10x_labels #mca_labels
blabels = ['FACS_'+x.split('.')[2] for x in tabula_data.index] + ['10x_'+mouse_dic['_'.join(x.split('_')[0:3])] for x in tabula_10x_data.index]
total_data = total_data.replace(np.NaN,0)
print('\nSummary')
print(total_data.shape)
print(set(labels))
print(set(labels_cell))
print(set(blabels))
#latent_space = TSNE(n_components=2)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
clustering_method = 'dbscan'
############################################
plt.figure(figsize=(16,40), dpi=300)
ax00 = plt.subplot2grid((10,4), (0,0))
ax10 = plt.subplot2grid((10,4), (0,1))
ax20 = plt.subplot2grid((10,4), (0,2))
ax30 = plt.subplot2grid((10,4), (0,3))
ax01 = plt.subplot2grid((10,4), (1,0))
ax11 = plt.subplot2grid((10,4), (1,1))
ax21 = plt.subplot2grid((10,4), (1,2))
ax31 = plt.subplot2grid((10,4), (1,3))
ax02 = plt.subplot2grid((10,4), (2,0))
ax12 = plt.subplot2grid((10,4), (2,1))
ax22 = plt.subplot2grid((10,4), (2,2))
ax32 = plt.subplot2grid((10,4), (2,3))
ax03 = plt.subplot2grid((10,4), (3,0))
ax13 = plt.subplot2grid((10,4), (3,1))
ax23 = plt.subplot2grid((10,4), (3,2))
ax33 = plt.subplot2grid((10,4), (3,3))
ax04 = plt.subplot2grid((10,4), (4,0))
ax14 = plt.subplot2grid((10,4), (4,1))
ax24 = plt.subplot2grid((10,4), (4,2))
ax34 = plt.subplot2grid((10,4), (4,3))
ax05 = plt.subplot2grid((10,4), (5,0))
ax15 = plt.subplot2grid((10,4), (5,1))
ax25 = plt.subplot2grid((10,4), (5,2))
ax35 = plt.subplot2grid((10,4), (5,3))
ax06 = plt.subplot2grid((10,4), (6,0))
ax16 = plt.subplot2grid((10,4), (6,1))
ax26 = plt.subplot2grid((10,4), (6,2))
ax36 = plt.subplot2grid((10,4), (6,3))
ax07 = plt.subplot2grid((10,4), (7,0))
ax17 = plt.subplot2grid((10,4), (7,1))
ax27 = plt.subplot2grid((10,4), (7,2))
ax37 = plt.subplot2grid((10,4), (7,3))
ax08 = plt.subplot2grid((10,4), (8,0))
ax18 = plt.subplot2grid((10,4), (8,1))
ax28 = plt.subplot2grid((10,4), (8,2))
ax38 = plt.subplot2grid((10,4), (8,3))
ax09 = plt.subplot2grid((10,4), (9,0))
ax19 = plt.subplot2grid((10,4), (9,1))
ax29 = plt.subplot2grid((10,4), (9,2))
ax39 = plt.subplot2grid((10,4), (9,3))
l = []
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_cp(total_data), \
ax00, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax05)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_cp(total_data)), \
ax10, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax15)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_total20000(df_cp(total_data)), \
ax20, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax25)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_total20000(df_cp(total_data))), \
ax30, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax35)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_cp(total_data)), \
ax01, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax06)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_cp(total_data))), \
ax11, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax16)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_total20000(df_cp(total_data))), \
ax21, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax26)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_total20000(df_cp(total_data)))), \
ax31, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax36)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_cp(total_data)), \
ax02, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax07)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_cp(total_data))), \
ax12, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax17)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_total20000(df_cp(total_data))), \
ax22, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax27)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_total20000(df_cp(total_data)))), \
ax32, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax37)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_cp(total_data)), \
ax03, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax08)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_cp(total_data))), \
ax13, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax18)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_total20000(df_cp(total_data))), \
ax23, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax28)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_total20000(df_cp(total_data)))), \
ax33, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax38)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_cp(total_data)), \
ax04, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax09)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_cp(total_data))), \
ax14, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax19)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_total20000(df_cp(total_data))), \
ax24, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax29)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_total20000(df_cp(total_data)))), \
ax34, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax39)
)
############################################
ax00.set_ylabel('raw' , fontsize=14)
ax01.set_ylabel('min-max norm' , fontsize=14)
ax02.set_ylabel('meansquare' , fontsize=14)
ax03.set_ylabel('l2 norm' , fontsize=14)
ax04.set_ylabel('z-score' , fontsize=14)
ax09.set_xlabel('raw', fontsize=13)
ax19.set_xlabel('log2', fontsize=13)
ax29.set_xlabel('total', fontsize=13)
ax39.set_xlabel('total_log2', fontsize=13)
ax34.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
ax39.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
['../data/tabular_muris/00_facs_raw_data/FACS/Kidney-counts.csv']
['../data/tabular_muris/01_droplet_raw_data/droplet/Kidney-10X_P7_5', '../data/tabular_muris/01_droplet_raw_data/droplet/Kidney-10X_P4_6', '../data/tabular_muris/01_droplet_raw_data/droplet/Kidney-10X_P4_5']
(865, 23433)
{'endothelial cell', 'fibroblast', 'kidney collecting duct cell', 'fenestrated cell', 'leukocyte', 'kidney tubule cell'}
(1264, 23433)
(2172, 23433)
(2782, 23433)
{'smooth muscle cell', 'endothelial cell', 'fibroblast', 'kidney collecting duct cell', 'macrophage', 'fenestrated cell', 'leukocyte', 'kidney tubule cell'}
Summary
(3298, 23432)
{'Kidney'}
{'smooth muscle cell', 'endothelial cell', 'fibroblast', 'kidney collecting duct cell', 'macrophage', 'fenestrated cell', 'leukocyte', 'kidney tubule cell'}
{'FACS_3_11_M', 'FACS_3_10_M', 'FACS_3_8_M', 'FACS_3_39_F', 'FACS_3_9_M', '10x_3-M-9', '10x_3-M-8', 'FACS_3_38_F', '10x_3-F-57'}
dbscan #cluster: (7, 0.12623650374585868, 0.080699906, 0.24413402191244266)
dbscan #cluster: (5, 0.47166238671454536, 0.7752027, 0.024973608637443124)
dbscan #cluster: (4, 0.44312538440072313, 0.4070545, 0.025210931932620903)
dbscan #cluster: (5, 0.3281223132500479, 0.85714304, 0.10039294100862846)
dbscan #cluster: (8, 0.24979040601498934, 0.3559891, 0.044099814287806974)
dbscan #cluster: (3, 0.7909050419027076, 0.7068792, 0.026166544735259162)
dbscan #cluster: (8, 0.24979040601498934, 0.3559891, 0.044099814287806974)
dbscan #cluster: (5, 0.31082515338060474, 0.81264585, 0.09208827240815087)
dbscan #cluster: (5, 0.5100168918234843, 0.2880439, 0.021654494016694812)
dbscan #cluster: (6, 0.4805003664378798, 0.8000584, 0.02476299249798733)
dbscan #cluster: (5, 0.2804692594573524, 0.6015514, 0.05722380764143755)
dbscan #cluster: (6, 0.44818591097182714, 0.62252635, 0.04027549900907555)
dbscan #cluster: (10, 0.15619888668442408, 0.37180686, 0.06599223367662448)
dbscan #cluster: (5, 0.48013375025897354, 0.7981096, 0.024740583351300513)
dbscan #cluster: (10, 0.15619888668442408, 0.37180686, 0.06599223367662448)
dbscan #cluster: (5, 0.44955374540504145, 0.6966349, 0.03989716305318338)
dbscan #cluster: (5, 0.47354662458627844, 0.39096877, 0.01630886412148596)
dbscan #cluster: (4, 0.454411549999143, 0.8590601, 0.02337878769396474)
dbscan #cluster: (5, 0.47354662458627844, 0.39096877, 0.01630886412148596)
dbscan #cluster: (6, 0.4479372334741431, 0.5861649, 0.039014844869572914)
<matplotlib.legend.Legend at 0x7fcfeff48880>
Muscle UMAP:DBSCAN results#
ORGAN = 'Muscle'
tabula_file_list = glob.glob("../data/tabular_muris/00_facs_raw_data/FACS/*"+ORGAN+"*.csv")
print(tabula_file_list)
tabula_10xfile_list = glob.glob("../data/tabular_muris/01_droplet_raw_data/droplet/*"+ORGAN+"*")
print(tabula_10xfile_list)
# Tabula data load
#
tabula_labels = []
tabula_data = pd.DataFrame()
for ff in tabula_file_list:
data = pd.read_csv(ff, sep=',', index_col=0, header=0)
data = data.transpose()
c = list(range(data.shape[0]))
tabula_data = pd.concat([tabula_data, data.iloc[c,]], axis=0)
tabula_labels += [ff.rstrip('-counts.csv').split('/')[-1]] * data.shape[0]
print(tabula_data.shape)
tabula_labels_set = set(tabula_labels)
# Tabula label load
#
annot_label = pd.read_csv('../data/tabular_muris/00_facs_raw_data/annotations_FACS.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_data.index)
m = annot_label.filter(com, axis=0)
tabula_data = tabula_data.filter(com, axis=0)
tabula_data = pd.concat([tabula_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue = tabula_data['tissue']
tabula_labels = tabula_data['cell_ontology_class']
tissue = tissue.values.tolist()
tabula_labels = tabula_labels.values.tolist()
tabula_data.pop('cell_ontology_class')
tabula_data.pop('tissue')
tabula_data.pop('zsGreen_transgene')
print(set(tabula_labels))
# Tabula 10x data load
#
tabula_10x_batches = []
tabula_10x_data = pd.DataFrame()
for ff in tabula_10xfile_list:
data = scanpy.read_10x_mtx(ff)
data = data.to_df()
prefix_cell = ff.split('-')[1]
data.index = [prefix_cell +'_'+x.split('-')[0] for x in data.index]
tabula_10x_data = pd.concat([tabula_10x_data, data], axis=0)
tabula_10x_batches += [ff.split('/')[-1].split('-')[1]] * data.shape[0]
print(tabula_10x_data.shape)
# Tabula 10x label load
#
annot_label = pd.read_csv('../data/tabular_muris/01_droplet_raw_data/annotations_droplet.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_10x_data.index)
m = annot_label.filter(com, axis=0)
tabula_10x_data = tabula_10x_data.filter(com, axis=0)
tabula_10x_data = pd.concat([tabula_10x_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue_10x = tabula_10x_data['tissue']
tabula_10x_labels = tabula_10x_data['cell_ontology_class']
tissue_10x = tissue_10x.values.tolist()
tabula_10x_labels = tabula_10x_labels.values.tolist()
tabula_10x_data.pop('cell_ontology_class')
tabula_10x_data.pop('tissue')
tabula_10x_data.pop('zsGreen_transgene')
print(set(tabula_10x_labels))
total_data = pd.concat([tabula_data, tabula_10x_data], axis=0)
labels = tissue + tissue_10x #mca_labels
labels_cell = tabula_labels + tabula_10x_labels #mca_labels
blabels = ['FACS_'+x.split('.')[2] for x in tabula_data.index] + ['10x_'+mouse_dic['_'.join(x.split('_')[0:3])] for x in tabula_10x_data.index]
total_data = total_data.replace(np.NaN,0)
print('\nSummary')
print(total_data.shape)
print(set(labels))
print(set(labels_cell))
print(set(blabels))
#latent_space = TSNE(n_components=2)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
clustering_method = 'dbscan'
############################################
plt.figure(figsize=(16,40), dpi=300)
ax00 = plt.subplot2grid((10,4), (0,0))
ax10 = plt.subplot2grid((10,4), (0,1))
ax20 = plt.subplot2grid((10,4), (0,2))
ax30 = plt.subplot2grid((10,4), (0,3))
ax01 = plt.subplot2grid((10,4), (1,0))
ax11 = plt.subplot2grid((10,4), (1,1))
ax21 = plt.subplot2grid((10,4), (1,2))
ax31 = plt.subplot2grid((10,4), (1,3))
ax02 = plt.subplot2grid((10,4), (2,0))
ax12 = plt.subplot2grid((10,4), (2,1))
ax22 = plt.subplot2grid((10,4), (2,2))
ax32 = plt.subplot2grid((10,4), (2,3))
ax03 = plt.subplot2grid((10,4), (3,0))
ax13 = plt.subplot2grid((10,4), (3,1))
ax23 = plt.subplot2grid((10,4), (3,2))
ax33 = plt.subplot2grid((10,4), (3,3))
ax04 = plt.subplot2grid((10,4), (4,0))
ax14 = plt.subplot2grid((10,4), (4,1))
ax24 = plt.subplot2grid((10,4), (4,2))
ax34 = plt.subplot2grid((10,4), (4,3))
ax05 = plt.subplot2grid((10,4), (5,0))
ax15 = plt.subplot2grid((10,4), (5,1))
ax25 = plt.subplot2grid((10,4), (5,2))
ax35 = plt.subplot2grid((10,4), (5,3))
ax06 = plt.subplot2grid((10,4), (6,0))
ax16 = plt.subplot2grid((10,4), (6,1))
ax26 = plt.subplot2grid((10,4), (6,2))
ax36 = plt.subplot2grid((10,4), (6,3))
ax07 = plt.subplot2grid((10,4), (7,0))
ax17 = plt.subplot2grid((10,4), (7,1))
ax27 = plt.subplot2grid((10,4), (7,2))
ax37 = plt.subplot2grid((10,4), (7,3))
ax08 = plt.subplot2grid((10,4), (8,0))
ax18 = plt.subplot2grid((10,4), (8,1))
ax28 = plt.subplot2grid((10,4), (8,2))
ax38 = plt.subplot2grid((10,4), (8,3))
ax09 = plt.subplot2grid((10,4), (9,0))
ax19 = plt.subplot2grid((10,4), (9,1))
ax29 = plt.subplot2grid((10,4), (9,2))
ax39 = plt.subplot2grid((10,4), (9,3))
l = []
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_cp(total_data), \
ax00, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax05)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_cp(total_data)), \
ax10, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax15)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_total20000(df_cp(total_data)), \
ax20, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax25)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_total20000(df_cp(total_data))), \
ax30, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax35)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_cp(total_data)), \
ax01, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax06)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_cp(total_data))), \
ax11, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax16)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_total20000(df_cp(total_data))), \
ax21, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax26)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_total20000(df_cp(total_data)))), \
ax31, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax36)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_cp(total_data)), \
ax02, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax07)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_cp(total_data))), \
ax12, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax17)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_total20000(df_cp(total_data))), \
ax22, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax27)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_total20000(df_cp(total_data)))), \
ax32, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax37)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_cp(total_data)), \
ax03, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax08)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_cp(total_data))), \
ax13, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax18)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_total20000(df_cp(total_data))), \
ax23, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax28)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_total20000(df_cp(total_data)))), \
ax33, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax38)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_cp(total_data)), \
ax04, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax09)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_cp(total_data))), \
ax14, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax19)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_total20000(df_cp(total_data))), \
ax24, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax29)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_total20000(df_cp(total_data)))), \
ax34, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax39)
)
############################################
ax00.set_ylabel('raw' , fontsize=14)
ax01.set_ylabel('min-max norm' , fontsize=14)
ax02.set_ylabel('meansquare' , fontsize=14)
ax03.set_ylabel('l2 norm' , fontsize=14)
ax04.set_ylabel('z-score' , fontsize=14)
ax09.set_xlabel('raw', fontsize=13)
ax19.set_xlabel('log2', fontsize=13)
ax29.set_xlabel('total', fontsize=13)
ax39.set_xlabel('total_log2', fontsize=13)
ax34.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
ax39.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
['../data/tabular_muris/00_facs_raw_data/FACS/Limb_Muscle-counts.csv']
['../data/tabular_muris/01_droplet_raw_data/droplet/Limb_Muscle-10X_P7_14', '../data/tabular_muris/01_droplet_raw_data/droplet/Limb_Muscle-10X_P7_15']
(1151, 23433)
{'B cell', 'macrophage', 'endothelial cell', 'T cell', 'mesenchymal stem cell', 'skeletal muscle satellite cell'}
(2242, 23433)
(4543, 23433)
{'macrophage', 'skeletal muscle satellite cell', 'endothelial cell', 'T cell', 'mesenchymal stem cell', 'B cell', 'chondroblast', 'unknown'}
Summary
(5603, 23432)
{'Limb_Muscle'}
{'B cell', 'macrophage', 'endothelial cell', 'T cell', 'mesenchymal stem cell', 'skeletal muscle satellite cell', 'chondroblast', 'unknown'}
{'FACS_3_11_M', 'FACS_3_39_F', 'FACS_3_8_M', 'FACS_3_10_M', '10x_3-F-56', 'FACS_3_9_M', 'FACS_3_38_F', '10x_3-F-57'}
dbscan #cluster: (5, 0.47470144542156245, 0.7031758, 0.15271185451788186)
dbscan #cluster: (10, 0.7460786423260025, 0.649035, 0.07401009098181224)
dbscan #cluster: (10, 0.6065257884637916, 0.6861861, 0.12703620817983635)
dbscan #cluster: (9, 0.5626666938434379, 0.46871236, 0.09444678505163939)
dbscan #cluster: (11, 0.5895015341535416, 0.50452536, 0.1274045450486752)
dbscan #cluster: (7, 0.8337078115994392, 0.719824, 0.02669082584497745)
dbscan #cluster: (11, 0.5895015341535416, 0.50452536, 0.1274045450486752)
dbscan #cluster: (8, 0.6979468718858985, 0.6908625, 0.045792472182891404)
dbscan #cluster: (11, 0.592737514101314, 0.60454917, 0.12972400604422693)
dbscan #cluster: (9, 0.8436572884277734, 0.6521653, 0.02536680779253134)
dbscan #cluster: (11, 0.592737514101314, 0.60454917, 0.12972400604422693)
dbscan #cluster: (10, 0.7194445226232223, 0.63199705, 0.08292107604534611)
dbscan #cluster: (10, 0.5832111553177975, 0.47992343, 0.13484160478563603)
dbscan #cluster: (9, 0.843736567650488, 0.6740441, 0.02537364962406117)
dbscan #cluster: (10, 0.5832111553177975, 0.47992343, 0.13484160478563603)
dbscan #cluster: (7, 0.6930446481345294, 0.72809803, 0.08243419625441836)
dbscan #cluster: (10, 0.5991286019110624, 0.66396046, 0.13051466706711923)
dbscan #cluster: (7, 0.8269748557950173, 0.8043667, 0.02869848585511388)
dbscan #cluster: (10, 0.5991286019110624, 0.66396046, 0.13051466706711923)
dbscan #cluster: (7, 0.6932769451447365, 0.71755224, 0.08175861467178835)
<matplotlib.legend.Legend at 0x7fcfd2614340>

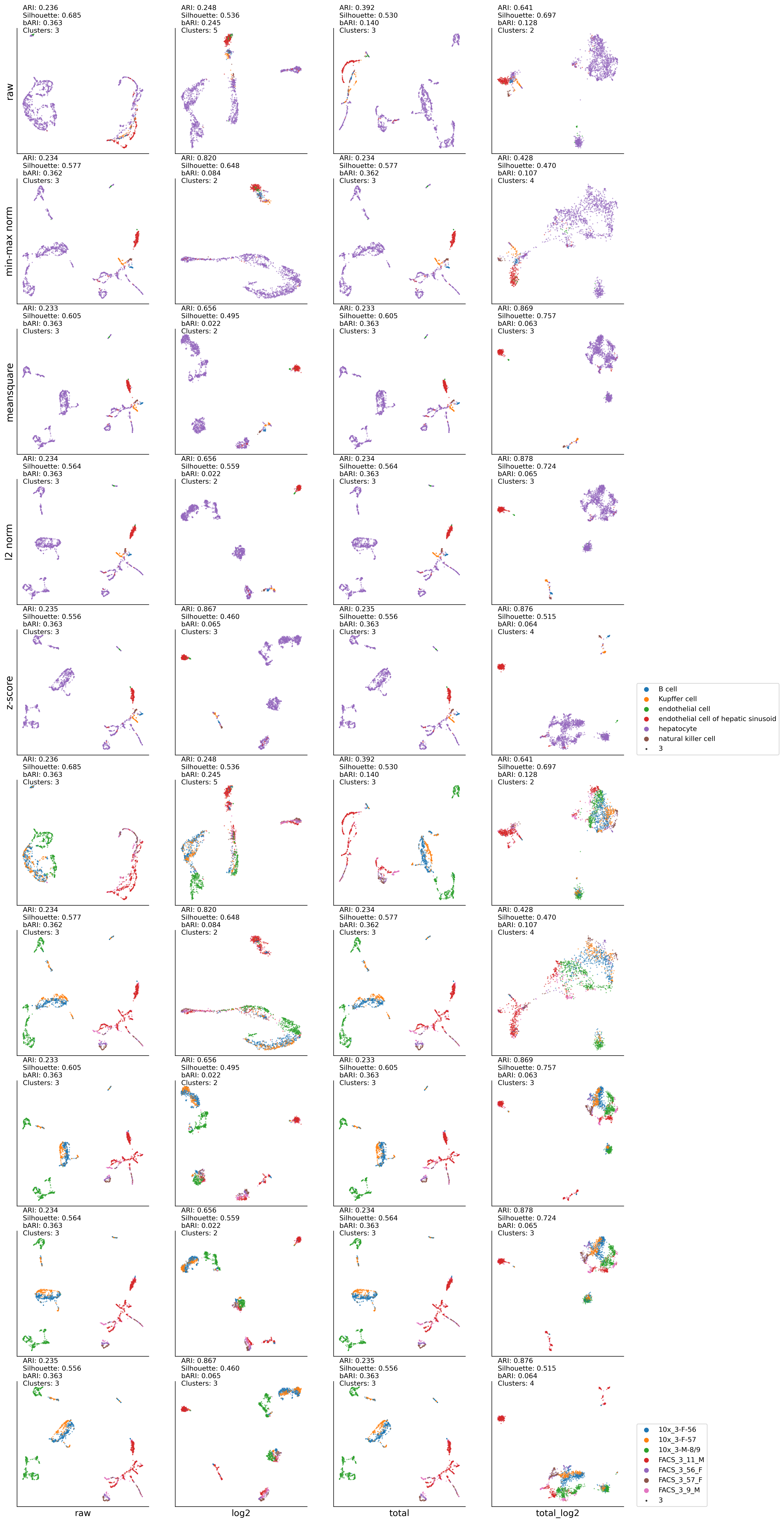
Liver UMAP:DBSCAN results#
ORGAN = 'Liver'
tabula_file_list = glob.glob("../data/tabular_muris/00_facs_raw_data/FACS/*"+ORGAN+"*.csv")
print(tabula_file_list)
tabula_10xfile_list = glob.glob("../data/tabular_muris/01_droplet_raw_data/droplet/*"+ORGAN+"*")
print(tabula_10xfile_list)
# Tabula data load
#
tabula_labels = []
tabula_data = pd.DataFrame()
for ff in tabula_file_list:
data = pd.read_csv(ff, sep=',', index_col=0, header=0)
data = data.transpose()
c = list(range(data.shape[0]))
tabula_data = pd.concat([tabula_data, data.iloc[c,]], axis=0)
tabula_labels += [ff.rstrip('-counts.csv').split('/')[-1]] * data.shape[0]
print(tabula_data.shape)
tabula_labels_set = set(tabula_labels)
# Tabula label load
#
annot_label = pd.read_csv('../data/tabular_muris/00_facs_raw_data/annotations_FACS.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_data.index)
m = annot_label.filter(com, axis=0)
tabula_data = tabula_data.filter(com, axis=0)
tabula_data = pd.concat([tabula_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue = tabula_data['tissue']
tabula_labels = tabula_data['cell_ontology_class']
tissue = tissue.values.tolist()
tabula_labels = tabula_labels.values.tolist()
tabula_data.pop('cell_ontology_class')
tabula_data.pop('tissue')
tabula_data.pop('zsGreen_transgene')
print(set(tabula_labels))
# Tabula 10x data load
#
tabula_10x_batches = []
tabula_10x_data = pd.DataFrame()
for ff in tabula_10xfile_list:
data = scanpy.read_10x_mtx(ff)
data = data.to_df()
prefix_cell = ff.split('-')[1]
data.index = [prefix_cell +'_'+x.split('-')[0] for x in data.index]
tabula_10x_data = pd.concat([tabula_10x_data, data], axis=0)
tabula_10x_batches += [ff.split('/')[-1].split('-')[1]] * data.shape[0]
print(tabula_10x_data.shape)
# Tabula 10x label load
#
annot_label = pd.read_csv('../data/tabular_muris/01_droplet_raw_data/annotations_droplet.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_10x_data.index)
m = annot_label.filter(com, axis=0)
tabula_10x_data = tabula_10x_data.filter(com, axis=0)
tabula_10x_data = pd.concat([tabula_10x_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue_10x = tabula_10x_data['tissue']
tabula_10x_labels = tabula_10x_data['cell_ontology_class']
tissue_10x = tissue_10x.values.tolist()
tabula_10x_labels = tabula_10x_labels.values.tolist()
tabula_10x_data.pop('cell_ontology_class')
tabula_10x_data.pop('tissue')
tabula_10x_data.pop('zsGreen_transgene')
print(set(tabula_10x_labels))
total_data = pd.concat([tabula_data, tabula_10x_data], axis=0)
labels = tissue + tissue_10x #mca_labels
labels_cell = tabula_labels + tabula_10x_labels #mca_labels
blabels = ['FACS_'+x.split('.')[2] for x in tabula_data.index] + ['10x_'+mouse_dic['_'.join(x.split('_')[0:3])] for x in tabula_10x_data.index]
total_data = total_data.replace(np.NaN,0)
print('\nSummary')
print(total_data.shape)
print(set(labels))
print(set(labels_cell))
print(set(blabels))
#latent_space = TSNE(n_components=2)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
clustering_method = 'dbscan'
############################################
plt.figure(figsize=(16,40), dpi=300)
ax00 = plt.subplot2grid((10,4), (0,0))
ax10 = plt.subplot2grid((10,4), (0,1))
ax20 = plt.subplot2grid((10,4), (0,2))
ax30 = plt.subplot2grid((10,4), (0,3))
ax01 = plt.subplot2grid((10,4), (1,0))
ax11 = plt.subplot2grid((10,4), (1,1))
ax21 = plt.subplot2grid((10,4), (1,2))
ax31 = plt.subplot2grid((10,4), (1,3))
ax02 = plt.subplot2grid((10,4), (2,0))
ax12 = plt.subplot2grid((10,4), (2,1))
ax22 = plt.subplot2grid((10,4), (2,2))
ax32 = plt.subplot2grid((10,4), (2,3))
ax03 = plt.subplot2grid((10,4), (3,0))
ax13 = plt.subplot2grid((10,4), (3,1))
ax23 = plt.subplot2grid((10,4), (3,2))
ax33 = plt.subplot2grid((10,4), (3,3))
ax04 = plt.subplot2grid((10,4), (4,0))
ax14 = plt.subplot2grid((10,4), (4,1))
ax24 = plt.subplot2grid((10,4), (4,2))
ax34 = plt.subplot2grid((10,4), (4,3))
ax05 = plt.subplot2grid((10,4), (5,0))
ax15 = plt.subplot2grid((10,4), (5,1))
ax25 = plt.subplot2grid((10,4), (5,2))
ax35 = plt.subplot2grid((10,4), (5,3))
ax06 = plt.subplot2grid((10,4), (6,0))
ax16 = plt.subplot2grid((10,4), (6,1))
ax26 = plt.subplot2grid((10,4), (6,2))
ax36 = plt.subplot2grid((10,4), (6,3))
ax07 = plt.subplot2grid((10,4), (7,0))
ax17 = plt.subplot2grid((10,4), (7,1))
ax27 = plt.subplot2grid((10,4), (7,2))
ax37 = plt.subplot2grid((10,4), (7,3))
ax08 = plt.subplot2grid((10,4), (8,0))
ax18 = plt.subplot2grid((10,4), (8,1))
ax28 = plt.subplot2grid((10,4), (8,2))
ax38 = plt.subplot2grid((10,4), (8,3))
ax09 = plt.subplot2grid((10,4), (9,0))
ax19 = plt.subplot2grid((10,4), (9,1))
ax29 = plt.subplot2grid((10,4), (9,2))
ax39 = plt.subplot2grid((10,4), (9,3))
l = []
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_cp(total_data), \
ax00, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax05)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_cp(total_data)), \
ax10, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax15)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_total20000(df_cp(total_data)), \
ax20, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax25)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_total20000(df_cp(total_data))), \
ax30, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax35)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_cp(total_data)), \
ax01, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax06)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_cp(total_data))), \
ax11, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax16)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_total20000(df_cp(total_data))), \
ax21, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax26)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_total20000(df_cp(total_data)))), \
ax31, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax36)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_cp(total_data)), \
ax02, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax07)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_cp(total_data))), \
ax12, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax17)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_total20000(df_cp(total_data))), \
ax22, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax27)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_total20000(df_cp(total_data)))), \
ax32, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax37)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_cp(total_data)), \
ax03, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax08)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_cp(total_data))), \
ax13, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax18)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_total20000(df_cp(total_data))), \
ax23, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax28)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_total20000(df_cp(total_data)))), \
ax33, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax38)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_cp(total_data)), \
ax04, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax09)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_cp(total_data))), \
ax14, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax19)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_total20000(df_cp(total_data))), \
ax24, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax29)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_total20000(df_cp(total_data)))), \
ax34, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax39)
)
############################################
ax00.set_ylabel('raw' , fontsize=14)
ax01.set_ylabel('min-max norm' , fontsize=14)
ax02.set_ylabel('meansquare' , fontsize=14)
ax03.set_ylabel('l2 norm' , fontsize=14)
ax04.set_ylabel('z-score' , fontsize=14)
ax09.set_xlabel('raw', fontsize=13)
ax19.set_xlabel('log2', fontsize=13)
ax29.set_xlabel('total', fontsize=13)
ax39.set_xlabel('total_log2', fontsize=13)
ax34.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
ax39.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
['../data/tabular_muris/00_facs_raw_data/FACS/Liver-counts.csv']
['../data/tabular_muris/01_droplet_raw_data/droplet/Liver-10X_P7_0', '../data/tabular_muris/01_droplet_raw_data/droplet/Liver-10X_P4_2', '../data/tabular_muris/01_droplet_raw_data/droplet/Liver-10X_P7_1']
(981, 23433)
{'hepatocyte', 'Kupffer cell', 'natural killer cell', 'B cell', 'endothelial cell of hepatic sinusoid'}
(596, 23433)
(1602, 23433)
(1924, 23433)
{'hepatocyte', 'endothelial cell'}
Summary
(1736, 23432)
{'Liver'}
{'hepatocyte', 'endothelial cell', 'Kupffer cell', 'natural killer cell', 'B cell', 'endothelial cell of hepatic sinusoid'}
{'FACS_3_11_M', '10x_3-M-8/9', '10x_3-F-56', 'FACS_3_56_F', 'FACS_3_57_F', 'FACS_3_9_M', '10x_3-F-57'}
dbscan #cluster: (3, 0.2360848659750596, 0.685125, 0.36328491741916763)
dbscan #cluster: (5, 0.24849027089847434, 0.5356156, 0.24470715281864047)
dbscan #cluster: (3, 0.39247285293394485, 0.52993506, 0.13977763103426788)
dbscan #cluster: (2, 0.6407463988820834, 0.6970445, 0.12802886377785994)
dbscan #cluster: (3, 0.2337845770088279, 0.57744396, 0.3619702681259615)
dbscan #cluster: (2, 0.8203122521927044, 0.6475501, 0.08443518288910089)
dbscan #cluster: (3, 0.2337845770088279, 0.57744396, 0.3619702681259615)
dbscan #cluster: (4, 0.428168370748511, 0.47037426, 0.10680557006436925)
dbscan #cluster: (3, 0.2326959126860678, 0.6051764, 0.362564467756735)
dbscan #cluster: (2, 0.656098978040263, 0.4951252, 0.022157832661340245)
dbscan #cluster: (3, 0.2326959126860678, 0.6051764, 0.362564467756735)
dbscan #cluster: (3, 0.8686335719253525, 0.757402, 0.06297171265627956)
dbscan #cluster: (3, 0.2337845770088279, 0.56352824, 0.36328875599104976)
dbscan #cluster: (2, 0.656098978040263, 0.5585497, 0.022157832661340245)
dbscan #cluster: (3, 0.2337845770088279, 0.56352824, 0.36328875599104976)
dbscan #cluster: (3, 0.8780325537940356, 0.72432345, 0.06535738455907514)
dbscan #cluster: (3, 0.23487545516762573, 0.5558794, 0.36269287885528373)
dbscan #cluster: (3, 0.8665180606818386, 0.46017078, 0.06540135822156201)
dbscan #cluster: (3, 0.23487545516762573, 0.5558794, 0.36269287885528373)
dbscan #cluster: (4, 0.8755455635621282, 0.5154985, 0.06407594949307682)
<matplotlib.legend.Legend at 0x7fcfe1b84e80>
Lung UMAP:DBSCAN results#
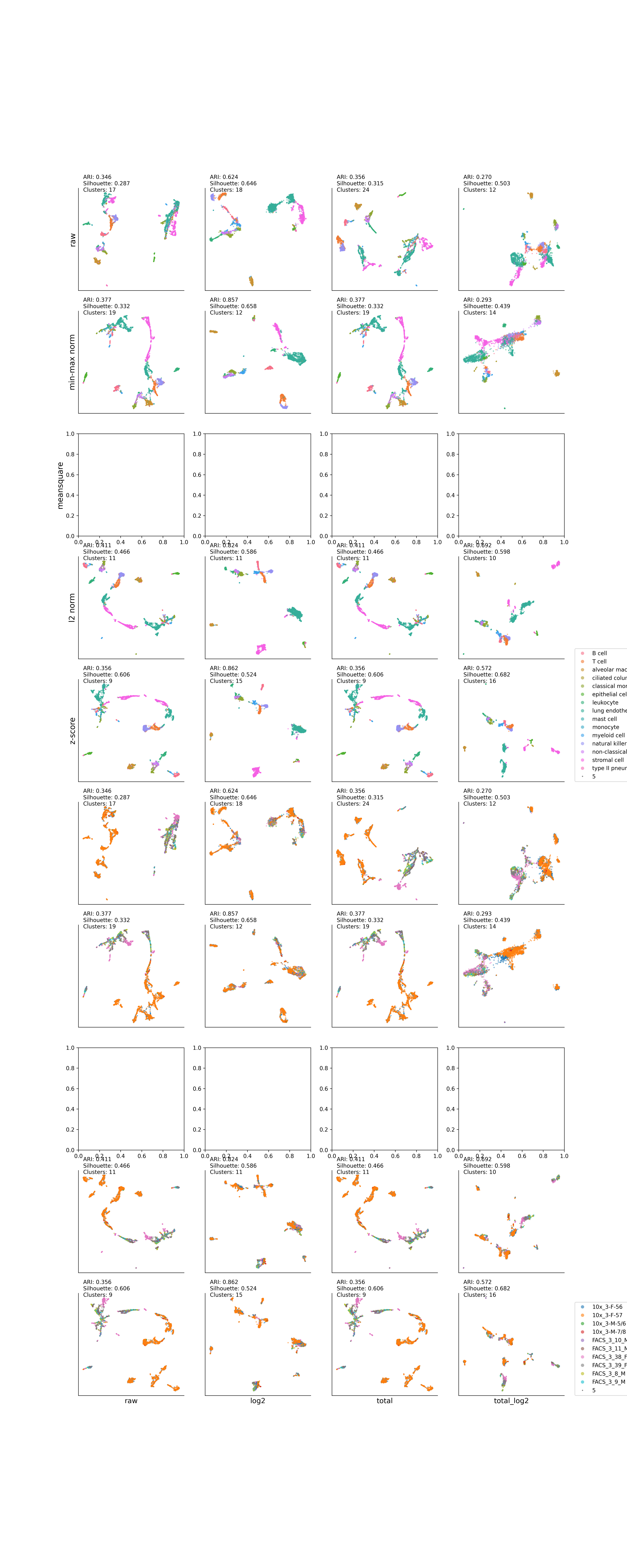
Mammary UMAP:DBSCAN results#
ORGAN = 'Mammary'
tabula_file_list = glob.glob("../data/tabular_muris/00_facs_raw_data/FACS/*"+ORGAN+"*.csv")
print(tabula_file_list)
tabula_10xfile_list = glob.glob("../data/tabular_muris/01_droplet_raw_data/droplet/*"+ORGAN+"*")
print(tabula_10xfile_list)
# Tabula data load
#
tabula_labels = []
tabula_data = pd.DataFrame()
for ff in tabula_file_list:
data = pd.read_csv(ff, sep=',', index_col=0, header=0)
data = data.transpose()
c = list(range(data.shape[0]))
tabula_data = pd.concat([tabula_data, data.iloc[c,]], axis=0)
tabula_labels += [ff.rstrip('-counts.csv').split('/')[-1]] * data.shape[0]
print(tabula_data.shape)
tabula_labels_set = set(tabula_labels)
# Tabula label load
#
annot_label = pd.read_csv('../data/tabular_muris/00_facs_raw_data/annotations_FACS.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_data.index)
m = annot_label.filter(com, axis=0)
tabula_data = tabula_data.filter(com, axis=0)
tabula_data = pd.concat([tabula_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue = tabula_data['tissue']
tabula_labels = tabula_data['cell_ontology_class']
tissue = tissue.values.tolist()
tabula_labels = tabula_labels.values.tolist()
tabula_data.pop('cell_ontology_class')
tabula_data.pop('tissue')
tabula_data.pop('zsGreen_transgene')
print(set(tabula_labels))
# Tabula 10x data load
#
tabula_10x_batches = []
tabula_10x_data = pd.DataFrame()
for ff in tabula_10xfile_list:
data = scanpy.read_10x_mtx(ff)
data = data.to_df()
prefix_cell = ff.split('-')[1]
data.index = [prefix_cell +'_'+x.split('-')[0] for x in data.index]
tabula_10x_data = pd.concat([tabula_10x_data, data], axis=0)
tabula_10x_batches += [ff.split('/')[-1].split('-')[1]] * data.shape[0]
print(tabula_10x_data.shape)
# Tabula 10x label load
#
annot_label = pd.read_csv('../data/tabular_muris/01_droplet_raw_data/annotations_droplet.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_10x_data.index)
m = annot_label.filter(com, axis=0)
tabula_10x_data = tabula_10x_data.filter(com, axis=0)
tabula_10x_data = pd.concat([tabula_10x_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue_10x = tabula_10x_data['tissue']
tabula_10x_labels = tabula_10x_data['cell_ontology_class']
tissue_10x = tissue_10x.values.tolist()
tabula_10x_labels = tabula_10x_labels.values.tolist()
tabula_10x_data.pop('cell_ontology_class')
tabula_10x_data.pop('tissue')
tabula_10x_data.pop('zsGreen_transgene')
print(set(tabula_10x_labels))
total_data = pd.concat([tabula_data, tabula_10x_data], axis=0)
labels = tissue + tissue_10x #mca_labels
labels_cell = tabula_labels + tabula_10x_labels #mca_labels
blabels = ['FACS_'+x.split('.')[2] for x in tabula_data.index] + ['10x_'+mouse_dic['_'.join(x.split('_')[0:3])] for x in tabula_10x_data.index]
total_data = total_data.replace(np.NaN,0)
print('\nSummary')
print(total_data.shape)
print(set(labels))
print(set(labels_cell))
print(set(blabels))
#latent_space = TSNE(n_components=2)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
clustering_method = 'dbscan'
############################################
plt.figure(figsize=(16,40), dpi=300)
ax00 = plt.subplot2grid((10,4), (0,0))
ax10 = plt.subplot2grid((10,4), (0,1))
ax20 = plt.subplot2grid((10,4), (0,2))
ax30 = plt.subplot2grid((10,4), (0,3))
ax01 = plt.subplot2grid((10,4), (1,0))
ax11 = plt.subplot2grid((10,4), (1,1))
ax21 = plt.subplot2grid((10,4), (1,2))
ax31 = plt.subplot2grid((10,4), (1,3))
ax02 = plt.subplot2grid((10,4), (2,0))
ax12 = plt.subplot2grid((10,4), (2,1))
ax22 = plt.subplot2grid((10,4), (2,2))
ax32 = plt.subplot2grid((10,4), (2,3))
ax03 = plt.subplot2grid((10,4), (3,0))
ax13 = plt.subplot2grid((10,4), (3,1))
ax23 = plt.subplot2grid((10,4), (3,2))
ax33 = plt.subplot2grid((10,4), (3,3))
ax04 = plt.subplot2grid((10,4), (4,0))
ax14 = plt.subplot2grid((10,4), (4,1))
ax24 = plt.subplot2grid((10,4), (4,2))
ax34 = plt.subplot2grid((10,4), (4,3))
ax05 = plt.subplot2grid((10,4), (5,0))
ax15 = plt.subplot2grid((10,4), (5,1))
ax25 = plt.subplot2grid((10,4), (5,2))
ax35 = plt.subplot2grid((10,4), (5,3))
ax06 = plt.subplot2grid((10,4), (6,0))
ax16 = plt.subplot2grid((10,4), (6,1))
ax26 = plt.subplot2grid((10,4), (6,2))
ax36 = plt.subplot2grid((10,4), (6,3))
ax07 = plt.subplot2grid((10,4), (7,0))
ax17 = plt.subplot2grid((10,4), (7,1))
ax27 = plt.subplot2grid((10,4), (7,2))
ax37 = plt.subplot2grid((10,4), (7,3))
ax08 = plt.subplot2grid((10,4), (8,0))
ax18 = plt.subplot2grid((10,4), (8,1))
ax28 = plt.subplot2grid((10,4), (8,2))
ax38 = plt.subplot2grid((10,4), (8,3))
ax09 = plt.subplot2grid((10,4), (9,0))
ax19 = plt.subplot2grid((10,4), (9,1))
ax29 = plt.subplot2grid((10,4), (9,2))
ax39 = plt.subplot2grid((10,4), (9,3))
l = []
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_cp(total_data), \
ax00, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax05)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_cp(total_data)), \
ax10, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax15)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_total20000(df_cp(total_data)), \
ax20, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax25)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_total20000(df_cp(total_data))), \
ax30, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax35)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_cp(total_data)), \
ax01, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax06)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_cp(total_data))), \
ax11, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax16)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_total20000(df_cp(total_data))), \
ax21, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax26)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_total20000(df_cp(total_data)))), \
ax31, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax36)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_cp(total_data)), \
ax02, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax07)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_cp(total_data))), \
ax12, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax17)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_total20000(df_cp(total_data))), \
ax22, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax27)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_total20000(df_cp(total_data)))), \
ax32, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax37)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_cp(total_data)), \
ax03, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax08)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_cp(total_data))), \
ax13, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax18)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_total20000(df_cp(total_data))), \
ax23, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax28)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_total20000(df_cp(total_data)))), \
ax33, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax38)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_cp(total_data)), \
ax04, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax09)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_cp(total_data))), \
ax14, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax19)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_total20000(df_cp(total_data))), \
ax24, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax29)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_total20000(df_cp(total_data)))), \
ax34, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax39)
)
############################################
ax00.set_ylabel('raw' , fontsize=14)
ax01.set_ylabel('min-max norm' , fontsize=14)
ax02.set_ylabel('meansquare' , fontsize=14)
ax03.set_ylabel('l2 norm' , fontsize=14)
ax04.set_ylabel('z-score' , fontsize=14)
ax09.set_xlabel('raw', fontsize=13)
ax19.set_xlabel('log2', fontsize=13)
ax29.set_xlabel('total', fontsize=13)
ax39.set_xlabel('total_log2', fontsize=13)
ax34.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
ax39.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
['../data/tabular_muris/00_facs_raw_data/FACS/Mammary_Gland-counts.csv']
['../data/tabular_muris/01_droplet_raw_data/droplet/Mammary_Gland-10X_P7_13', '../data/tabular_muris/01_droplet_raw_data/droplet/Mammary_Gland-10X_P7_12']
(2663, 23433)
{'basal cell', 'stromal cell', 'luminal epithelial cell of mammary gland', 'endothelial cell'}
(2543, 23433)
(4481, 23433)
{'macrophage', 'B cell', 'endothelial cell', 'basal cell', 'T cell', 'stromal cell', 'luminal cell of lactiferous duct'}
Summary
(6785, 23432)
{'Mammary_Gland'}
{'macrophage', 'B cell', 'endothelial cell', 'basal cell', 'luminal epithelial cell of mammary gland', 'T cell', 'stromal cell', 'luminal cell of lactiferous duct'}
{'FACS_3_56_F', '10x_3-F-57', 'FACS_3_39_F', 'FACS_3_57_F', 'FACS_3_38_F', '10x_3-F-56'}
dbscan #cluster: (7, 0.39062471994657316, 0.17230041, 0.30246951107907755)
dbscan #cluster: (7, 0.69986416518005, 0.7918188, 0.1163610451599924)
dbscan #cluster: (9, 0.6958207346525456, 0.7319836, 0.1751627504462043)
dbscan #cluster: (11, 0.5970614469708084, 0.73380995, 0.26885651954414697)
dbscan #cluster: (7, 0.5205937144232875, 0.5813926, 0.2752801857877461)
dbscan #cluster: (7, 0.8842309229965729, 0.7101749, 0.10396534247008377)
dbscan #cluster: (7, 0.5205937144232875, 0.5813926, 0.2752801857877461)
dbscan #cluster: (6, 0.6209786324546772, 0.8352822, 0.26373922535224514)
dbscan #cluster: (8, 0.6861233788627884, 0.7396736, 0.18429687251197607)
dbscan #cluster: (8, 0.8965133052875178, 0.80671537, 0.09409891296042187)
dbscan #cluster: (8, 0.6861233788627884, 0.7396736, 0.18429687251197607)
dbscan #cluster: (6, 0.619110213373637, 0.84141934, 0.23626699268654122)
dbscan #cluster: (9, 0.6722147042940677, 0.71142125, 0.18404084425242173)
dbscan #cluster: (8, 0.896236616221339, 0.81834483, 0.09410604570392679)
dbscan #cluster: (9, 0.6722147042940677, 0.71142125, 0.18404084425242173)
dbscan #cluster: (6, 0.6190557764183131, 0.83971745, 0.2365533771183592)
dbscan #cluster: (10, 0.7185708058656488, 0.6834063, 0.18925381784471867)
dbscan #cluster: (7, 0.8948420041226876, 0.83494407, 0.09499879705837777)
dbscan #cluster: (8, 0.6869342072101516, 0.7773397, 0.18461045655090452)
dbscan #cluster: (7, 0.6417483844000249, 0.85020894, 0.25536674587264235)
<matplotlib.legend.Legend at 0x7f4f3ebaf130>
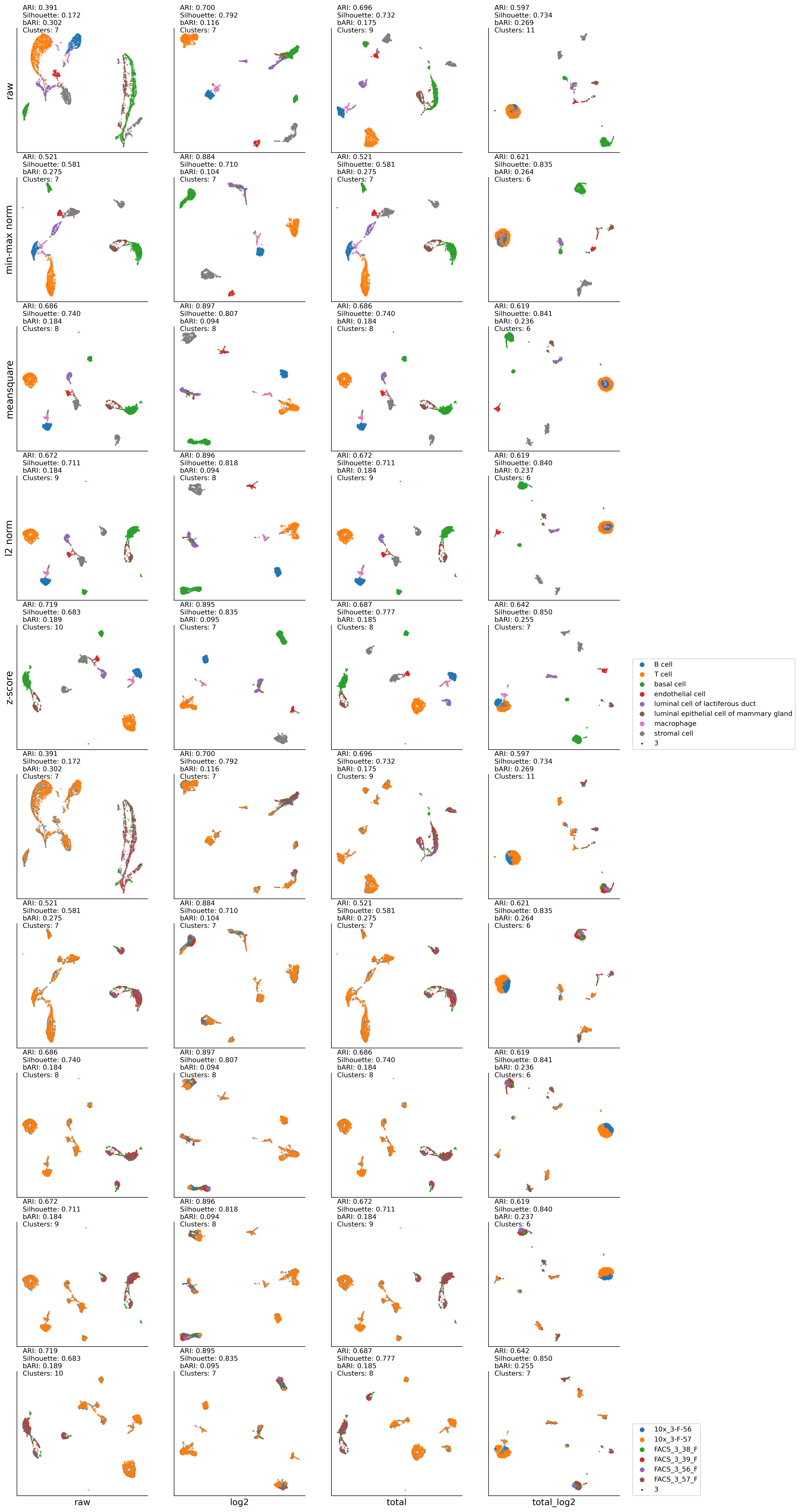
Marrow UMAP:DBSCAN results#
ORGAN = 'Marrow'
tabula_file_list = glob.glob("./dataset/00_facs_raw_data/FACS/*"+ORGAN+"*.csv")
print(tabula_file_list)
tabula_10xfile_list = glob.glob("./dataset/01_droplet_raw_data/droplet/*"+ORGAN+"*")
print(tabula_10xfile_list)
# Tabula data load
#
tabula_labels = []
tabula_data = pd.DataFrame()
for ff in tabula_file_list:
data = pd.read_csv(ff, sep=',', index_col=0, header=0)
data = data.transpose()
c = list(range(data.shape[0]))
random.shuffle(c)
# c = c[:sample_per_class]
tabula_data = pd.concat([tabula_data, data.iloc[c,]], axis=0)
tabula_labels += [ff.rstrip('-counts.csv').split('/')[-1]] * data.shape[0]
print(tabula_data.shape)
tabula_labels_set = set(tabula_labels)
# Tabula label load
#
annot_label = pd.read_csv('./dataset/00_facs_raw_data/annotations_FACS.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_data.index)
m = annot_label.filter(com, axis=0)
tabula_data = tabula_data.filter(com, axis=0)
tabula_data = pd.concat([tabula_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue = tabula_data['tissue']
tabula_labels = tabula_data['cell_ontology_class']
tissue = tissue.values.tolist()
tabula_labels = tabula_labels.values.tolist()
tabula_data.pop('cell_ontology_class')
tabula_data.pop('tissue')
tabula_data.pop('zsGreen_transgene')
print(set(tabula_labels))
# Tabula 10x data load
#
#sample_per_class = 2000
tabula_10x_batches = []
tabula_10x_data = pd.DataFrame()
for ff in tabula_10xfile_list:
data = scanpy.read_10x_mtx(ff)
c = list(range(len(data.obs)))
random.shuffle(c)
# c = c[:sample_per_class]
data = data[c,]
data = data.to_df()
prefix_cell = ff.split('-')[1]
data.index = [prefix_cell +'_'+x.split('-')[0] for x in data.index]
tabula_10x_data = pd.concat([tabula_10x_data, data], axis=0)
tabula_10x_batches += [ff.split('/')[-1].split('-')[1]] * data.shape[0]
print(tabula_10x_data.shape)
# Tabula 10x label load
#
annot_label = pd.read_csv('./dataset/01_droplet_raw_data/annotations_droplet.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_10x_data.index)
m = annot_label.filter(com, axis=0)
tabula_10x_data = tabula_10x_data.filter(com, axis=0)
tabula_10x_data = pd.concat([tabula_10x_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue_10x = tabula_10x_data['tissue']
tabula_10x_labels = tabula_10x_data['cell_ontology_class']
tissue_10x = tissue_10x.values.tolist()
tabula_10x_labels = tabula_10x_labels.values.tolist()
tabula_10x_data.pop('cell_ontology_class')
tabula_10x_data.pop('tissue')
tabula_10x_data.pop('zsGreen_transgene')
print(set(tabula_10x_labels))
total_data = pd.concat([tabula_data, tabula_10x_data], axis=0)
labels = tissue + tissue_10x #mca_labels
labels_cell = tabula_labels + tabula_10x_labels #mca_labels
blabels = ['FACS_'+x.split('.')[2] for x in tabula_data.index] + ['10x_'+mouse_dic['_'.join(x.split('_')[0:3])] for x in tabula_10x_data.index]
total_data = total_data.replace(np.NaN,0)
print('\nSummary')
print(total_data.shape)
print(set(labels))
print(set(labels_cell))
print(set(blabels))
#latent_space = TSNE(n_components=2)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
clustering_method = 'dbscan'
############################################
plt.figure(figsize=(16,40), dpi=300)
ax00 = plt.subplot2grid((10,4), (0,0))
ax10 = plt.subplot2grid((10,4), (0,1))
ax20 = plt.subplot2grid((10,4), (0,2))
ax30 = plt.subplot2grid((10,4), (0,3))
ax01 = plt.subplot2grid((10,4), (1,0))
ax11 = plt.subplot2grid((10,4), (1,1))
ax21 = plt.subplot2grid((10,4), (1,2))
ax31 = plt.subplot2grid((10,4), (1,3))
ax02 = plt.subplot2grid((10,4), (2,0))
ax12 = plt.subplot2grid((10,4), (2,1))
ax22 = plt.subplot2grid((10,4), (2,2))
ax32 = plt.subplot2grid((10,4), (2,3))
ax03 = plt.subplot2grid((10,4), (3,0))
ax13 = plt.subplot2grid((10,4), (3,1))
ax23 = plt.subplot2grid((10,4), (3,2))
ax33 = plt.subplot2grid((10,4), (3,3))
ax04 = plt.subplot2grid((10,4), (4,0))
ax14 = plt.subplot2grid((10,4), (4,1))
ax24 = plt.subplot2grid((10,4), (4,2))
ax34 = plt.subplot2grid((10,4), (4,3))
ax05 = plt.subplot2grid((10,4), (5,0))
ax15 = plt.subplot2grid((10,4), (5,1))
ax25 = plt.subplot2grid((10,4), (5,2))
ax35 = plt.subplot2grid((10,4), (5,3))
ax06 = plt.subplot2grid((10,4), (6,0))
ax16 = plt.subplot2grid((10,4), (6,1))
ax26 = plt.subplot2grid((10,4), (6,2))
ax36 = plt.subplot2grid((10,4), (6,3))
ax07 = plt.subplot2grid((10,4), (7,0))
ax17 = plt.subplot2grid((10,4), (7,1))
ax27 = plt.subplot2grid((10,4), (7,2))
ax37 = plt.subplot2grid((10,4), (7,3))
ax08 = plt.subplot2grid((10,4), (8,0))
ax18 = plt.subplot2grid((10,4), (8,1))
ax28 = plt.subplot2grid((10,4), (8,2))
ax38 = plt.subplot2grid((10,4), (8,3))
ax09 = plt.subplot2grid((10,4), (9,0))
ax19 = plt.subplot2grid((10,4), (9,1))
ax29 = plt.subplot2grid((10,4), (9,2))
ax39 = plt.subplot2grid((10,4), (9,3))
l = []
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_cp(total_data), \
ax00, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax05)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_cp(total_data)), \
ax10, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax15)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_total20000(df_cp(total_data)), \
ax20, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax25)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_total20000(df_cp(total_data))), \
ax30, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax35)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_cp(total_data)), \
ax01, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax06)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_cp(total_data))), \
ax11, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax16)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_total20000(df_cp(total_data))), \
ax21, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax26)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_total20000(df_cp(total_data)))), \
ax31, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax36)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_cp(total_data)), \
ax02, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax07)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_cp(total_data))), \
ax12, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax17)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_total20000(df_cp(total_data))), \
ax22, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax27)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_total20000(df_cp(total_data)))), \
ax32, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax37)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_cp(total_data)), \
ax03, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax08)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_cp(total_data))), \
ax13, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax18)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_total20000(df_cp(total_data))), \
ax23, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax28)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_total20000(df_cp(total_data)))), \
ax33, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax38)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_cp(total_data)), \
ax04, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax09)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_cp(total_data))), \
ax14, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax19)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_total20000(df_cp(total_data))), \
ax24, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax29)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_total20000(df_cp(total_data)))), \
ax34, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax39)
)
############################################
ax00.set_ylabel('raw' , fontsize=14)
ax01.set_ylabel('min-max norm' , fontsize=14)
ax02.set_ylabel('meansquare' , fontsize=14)
ax03.set_ylabel('l2 norm' , fontsize=14)
ax04.set_ylabel('z-score' , fontsize=14)
ax09.set_xlabel('raw', fontsize=13)
ax19.set_xlabel('log2', fontsize=13)
ax29.set_xlabel('total', fontsize=13)
ax39.set_xlabel('total_log2', fontsize=13)
ax34.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
ax39.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
['./dataset/00_facs_raw_data/FACS/Marrow-counts.csv']
['./dataset/01_droplet_raw_data/droplet/Marrow-10X_P7_2', './dataset/01_droplet_raw_data/droplet/Marrow-10X_P7_3']
(5355, 23433)
{'monocyte', 'neutrophil', 'natural killer cell', 'T cell', 'granulocyte', 'Fraction A pre-pro B cell', 'hematopoietic stem cell', 'B cell'}
(1981, 23433)
(4112, 23433)
{'monocyte', 'macrophage', 'erythrocyte', 'T cell', 'granulocyte', 'Fraction A pre-pro B cell', 'hematopoietic stem cell', 'B cell'}
Summary
(8549, 23432)
{'Marrow'}
{'monocyte', 'macrophage', 'erythrocyte', 'neutrophil', 'natural killer cell', 'T cell', 'granulocyte', 'Fraction A pre-pro B cell', 'hematopoietic stem cell', 'B cell'}
{'10x_3-F-56', '10x_3-F-57', 'FACS_3_38_F', 'FACS_3_10_M', 'FACS_3_8_M', 'FACS_3_9_M', 'FACS_3_39_F'}
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/opt/conda/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
<matplotlib.legend.Legend at 0x7fe3abe6b8e0>

Spleen UMAP:DBSCAN results#
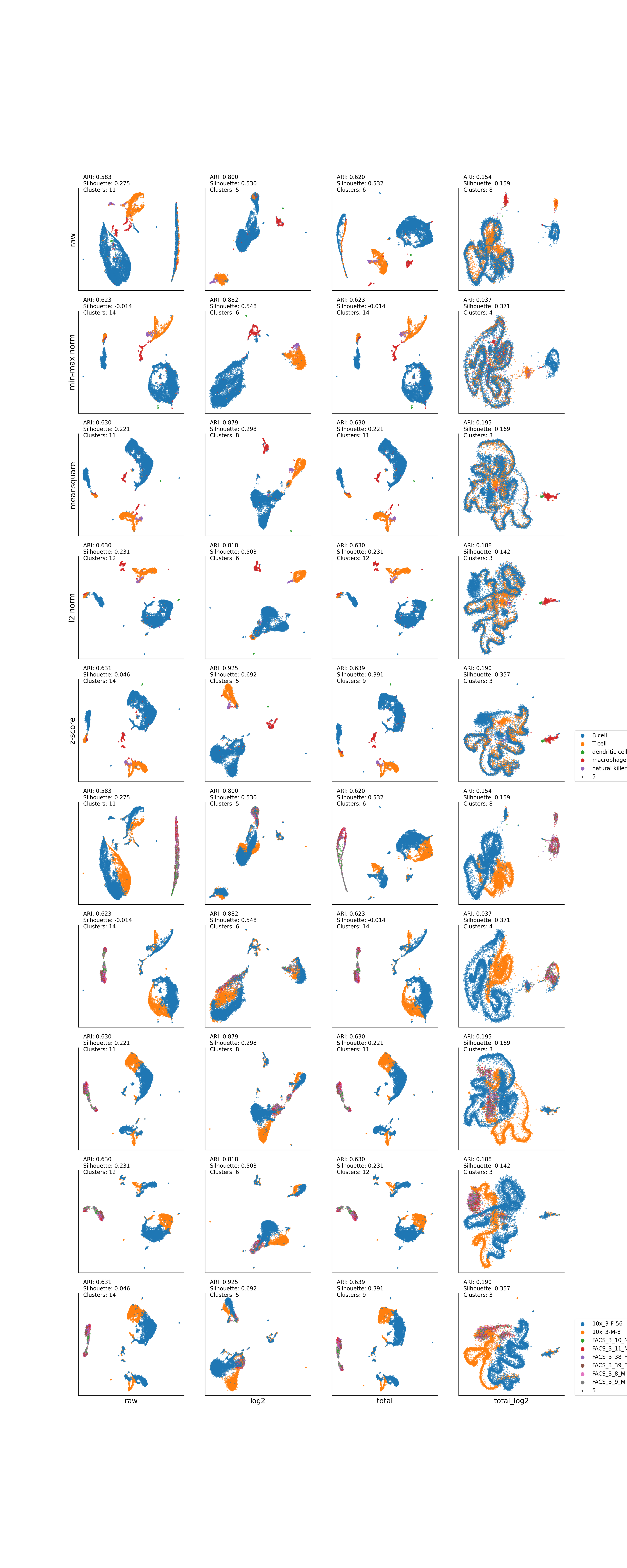
Thymus UMAP:DBSCAN results#
ORGAN = 'Thymus'
tabula_file_list = glob.glob("../data/tabular_muris/00_facs_raw_data/FACS/*"+ORGAN+"*.csv")
print(tabula_file_list)
tabula_10xfile_list = glob.glob("../data/tabular_muris/01_droplet_raw_data/droplet/*"+ORGAN+"*")
print(tabula_10xfile_list)
# Tabula data load
#
tabula_labels = []
tabula_data = pd.DataFrame()
for ff in tabula_file_list:
data = pd.read_csv(ff, sep=',', index_col=0, header=0)
data = data.transpose()
c = list(range(data.shape[0]))
tabula_data = pd.concat([tabula_data, data.iloc[c,]], axis=0)
tabula_labels += [ff.rstrip('-counts.csv').split('/')[-1]] * data.shape[0]
print(tabula_data.shape)
tabula_labels_set = set(tabula_labels)
# Tabula label load
#
annot_label = pd.read_csv('../data/tabular_muris/00_facs_raw_data/annotations_FACS.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_data.index)
m = annot_label.filter(com, axis=0)
tabula_data = tabula_data.filter(com, axis=0)
tabula_data = pd.concat([tabula_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue = tabula_data['tissue']
tabula_labels = tabula_data['cell_ontology_class']
tissue = tissue.values.tolist()
tabula_labels = tabula_labels.values.tolist()
tabula_data.pop('cell_ontology_class')
tabula_data.pop('tissue')
tabula_data.pop('zsGreen_transgene')
print(set(tabula_labels))
# Tabula 10x data load
#
tabula_10x_batches = []
tabula_10x_data = pd.DataFrame()
for ff in tabula_10xfile_list:
data = scanpy.read_10x_mtx(ff)
data = data.to_df()
prefix_cell = ff.split('-')[1]
data.index = [prefix_cell +'_'+x.split('-')[0] for x in data.index]
tabula_10x_data = pd.concat([tabula_10x_data, data], axis=0)
tabula_10x_batches += [ff.split('/')[-1].split('-')[1]] * data.shape[0]
print(tabula_10x_data.shape)
# Tabula 10x label load
#
annot_label = pd.read_csv('../data/tabular_muris/01_droplet_raw_data/annotations_droplet.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(tabula_10x_data.index)
m = annot_label.filter(com, axis=0)
tabula_10x_data = tabula_10x_data.filter(com, axis=0)
tabula_10x_data = pd.concat([tabula_10x_data, m[['tissue','cell_ontology_class']]],axis =1)
tissue_10x = tabula_10x_data['tissue']
tabula_10x_labels = tabula_10x_data['cell_ontology_class']
tissue_10x = tissue_10x.values.tolist()
tabula_10x_labels = tabula_10x_labels.values.tolist()
tabula_10x_data.pop('cell_ontology_class')
tabula_10x_data.pop('tissue')
tabula_10x_data.pop('zsGreen_transgene')
print(set(tabula_10x_labels))
total_data = pd.concat([tabula_data, tabula_10x_data], axis=0)
labels = tissue + tissue_10x #mca_labels
labels_cell = tabula_labels + tabula_10x_labels #mca_labels
blabels = ['FACS_'+x.split('.')[2] for x in tabula_data.index] + ['10x_'+mouse_dic['_'.join(x.split('_')[0:3])] for x in tabula_10x_data.index]
total_data = total_data.replace(np.NaN,0)
print('\nSummary')
print(total_data.shape)
print(set(labels))
print(set(labels_cell))
print(set(blabels))
#latent_space = TSNE(n_components=2)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
clustering_method = 'dbscan'
############################################
plt.figure(figsize=(16,40), dpi=300)
ax00 = plt.subplot2grid((10,4), (0,0))
ax10 = plt.subplot2grid((10,4), (0,1))
ax20 = plt.subplot2grid((10,4), (0,2))
ax30 = plt.subplot2grid((10,4), (0,3))
ax01 = plt.subplot2grid((10,4), (1,0))
ax11 = plt.subplot2grid((10,4), (1,1))
ax21 = plt.subplot2grid((10,4), (1,2))
ax31 = plt.subplot2grid((10,4), (1,3))
ax02 = plt.subplot2grid((10,4), (2,0))
ax12 = plt.subplot2grid((10,4), (2,1))
ax22 = plt.subplot2grid((10,4), (2,2))
ax32 = plt.subplot2grid((10,4), (2,3))
ax03 = plt.subplot2grid((10,4), (3,0))
ax13 = plt.subplot2grid((10,4), (3,1))
ax23 = plt.subplot2grid((10,4), (3,2))
ax33 = plt.subplot2grid((10,4), (3,3))
ax04 = plt.subplot2grid((10,4), (4,0))
ax14 = plt.subplot2grid((10,4), (4,1))
ax24 = plt.subplot2grid((10,4), (4,2))
ax34 = plt.subplot2grid((10,4), (4,3))
ax05 = plt.subplot2grid((10,4), (5,0))
ax15 = plt.subplot2grid((10,4), (5,1))
ax25 = plt.subplot2grid((10,4), (5,2))
ax35 = plt.subplot2grid((10,4), (5,3))
ax06 = plt.subplot2grid((10,4), (6,0))
ax16 = plt.subplot2grid((10,4), (6,1))
ax26 = plt.subplot2grid((10,4), (6,2))
ax36 = plt.subplot2grid((10,4), (6,3))
ax07 = plt.subplot2grid((10,4), (7,0))
ax17 = plt.subplot2grid((10,4), (7,1))
ax27 = plt.subplot2grid((10,4), (7,2))
ax37 = plt.subplot2grid((10,4), (7,3))
ax08 = plt.subplot2grid((10,4), (8,0))
ax18 = plt.subplot2grid((10,4), (8,1))
ax28 = plt.subplot2grid((10,4), (8,2))
ax38 = plt.subplot2grid((10,4), (8,3))
ax09 = plt.subplot2grid((10,4), (9,0))
ax19 = plt.subplot2grid((10,4), (9,1))
ax29 = plt.subplot2grid((10,4), (9,2))
ax39 = plt.subplot2grid((10,4), (9,3))
l = []
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_cp(total_data), \
ax00, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax05)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_cp(total_data)), \
ax10, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax15)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_total20000(df_cp(total_data)), \
ax20, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax25)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_total20000(df_cp(total_data))), \
ax30, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax35)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_cp(total_data)), \
ax01, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax06)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_cp(total_data))), \
ax11, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax16)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_total20000(df_cp(total_data))), \
ax21, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax26)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_total20000(df_cp(total_data)))), \
ax31, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax36)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_cp(total_data)), \
ax02, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax07)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_cp(total_data))), \
ax12, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax17)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_total20000(df_cp(total_data))), \
ax22, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax27)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_total20000(df_cp(total_data)))), \
ax32, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax37)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_cp(total_data)), \
ax03, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax08)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_cp(total_data))), \
ax13, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax18)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_total20000(df_cp(total_data))), \
ax23, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax28)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_total20000(df_cp(total_data)))), \
ax33, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax38)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_cp(total_data)), \
ax04, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax09)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_cp(total_data))), \
ax14, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax19)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_total20000(df_cp(total_data))), \
ax24, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax29)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_total20000(df_cp(total_data)))), \
ax34, labels_cell, latent_space, clustering_method, blabels=blabels, b_ax=ax39)
)
############################################
ax00.set_ylabel('raw' , fontsize=14)
ax01.set_ylabel('min-max norm' , fontsize=14)
ax02.set_ylabel('meansquare' , fontsize=14)
ax03.set_ylabel('l2 norm' , fontsize=14)
ax04.set_ylabel('z-score' , fontsize=14)
ax09.set_xlabel('raw', fontsize=13)
ax19.set_xlabel('log2', fontsize=13)
ax29.set_xlabel('total', fontsize=13)
ax39.set_xlabel('total_log2', fontsize=13)
ax34.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
ax39.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
['../data/tabular_muris/00_facs_raw_data/FACS/Thymus-counts.csv']
['../data/tabular_muris/01_droplet_raw_data/droplet/Thymus-10X_P7_11']
(1580, 23433)
{'T cell', 'mesenchymal stem cell'}
(1431, 23433)
{'T cell', 'stromal cell'}
Summary
(2712, 23432)
{'Thymus'}
{'T cell', 'stromal cell', 'mesenchymal stem cell'}
{'FACS_3_39_F', '10x_3-F-56', 'FACS_3_8_M', 'FACS_3_38_F', 'FACS_3_9_M', 'FACS_3_11_M'}
dbscan #cluster: (3, 0.006578671453967053, 0.53926647, 0.34440518832008665)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (5, 0.03357352211175827, 0.46550608, 0.42211129625531024)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (3, 0.0016585593776565599, 0.3619248, 0.6332560940023169)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (2, 0.11694366395833133, 0.12602337, -0.004364810887809986)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (3, 0.5361015482417778, -0.23462464, 0.004409193200951024)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (2, 0.11694366395833133, 0.17210685, -0.004364810887809986)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (2, 0.5451234144079106, 0.053586043, 0.0028120777420688305)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (3, 0.5322312214905789, -0.21933818, 0.004399205252496503)
<matplotlib.legend.Legend at 0x7f68d1c360a0>
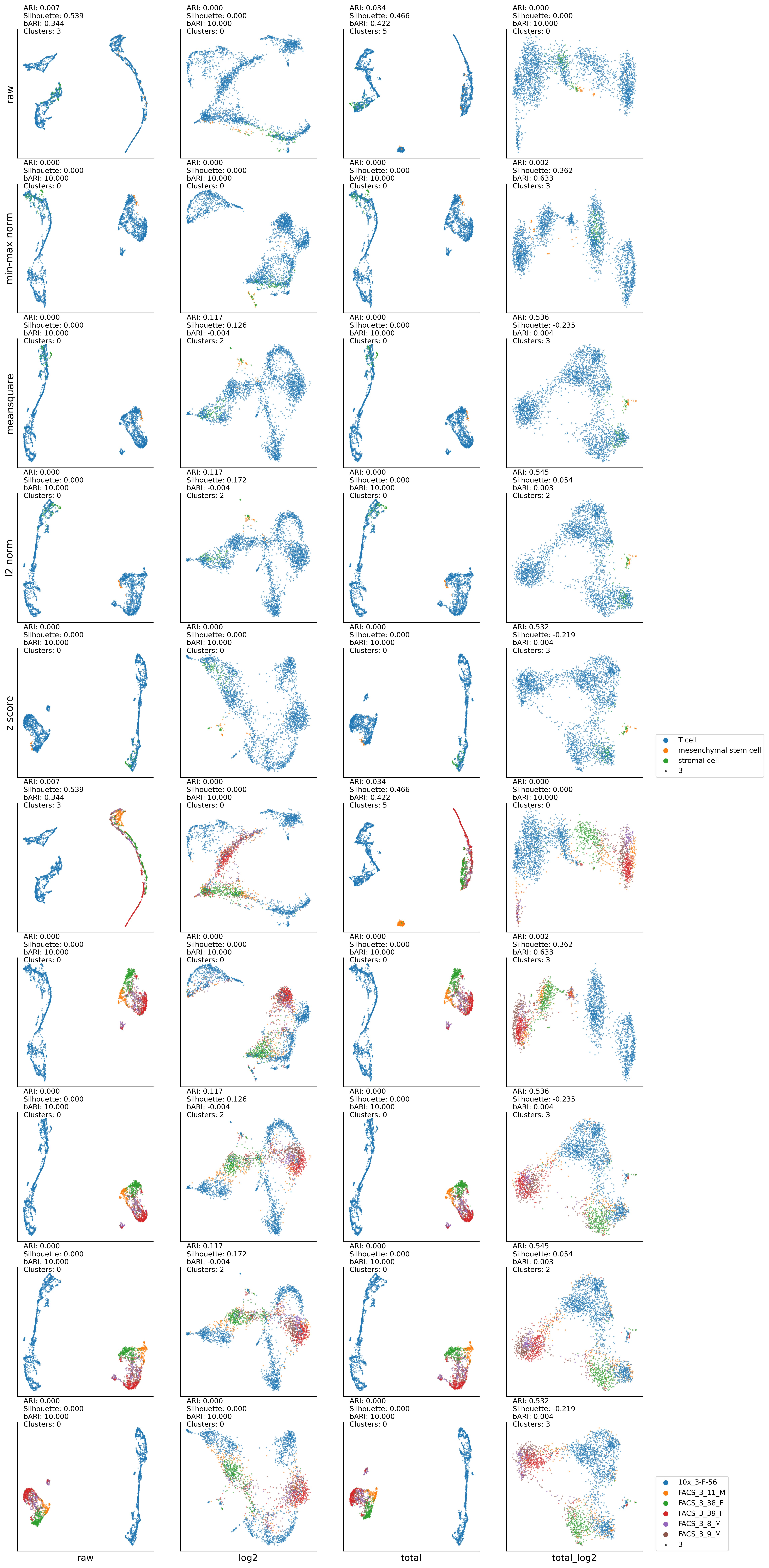
Tongue UMAP:DBSCAN results#
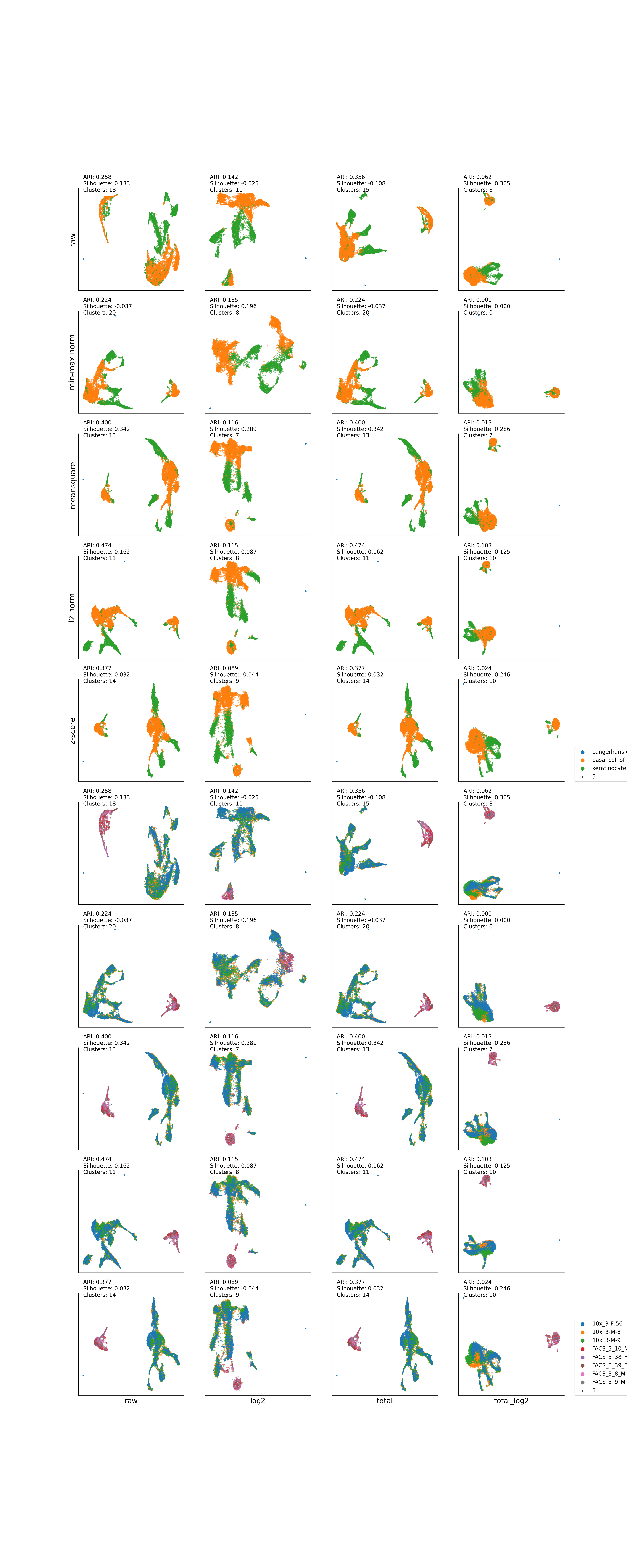
Trachea UMAP:DBSCAN results#