1. Conventional analysis of a scRNA-seq data#
%pip install umap
%pip install scanpy
%pip install 'scanpy[leiden]'
%pip install louvain
import warnings
warnings.filterwarnings('ignore')
import sys
sys.path.append('./src')
import utils as my_u
from utils import data_transformation
from utils import clustering
from utils import h5_data_loader
#import scanpy.api as sc
import anndata as ad
import scanpy as sc
import logging
import numpy as np
from sklearn.decomposition import PCA
from sklearn import preprocessing
from sklearn.metrics.cluster import normalized_mutual_info_score
from sklearn.metrics.cluster import adjusted_rand_score
import umap
Baron: Scanpy-HVG#
datasets = ['./dataset/baron_sc.h5']
label_filter = ['epsilon', 'alpha', 'beta', 'duct', 'activated', 'schwann', 'gamma', 'quiescent', 'delta', 'macrophage', 'endothelial', 'acinar', 'mast']
X_, y_, b_, file_names = h5_data_loader(datasets, label_filter)
logging.info(f'Data loaded. {datasets}')
adata = ad.AnnData(X_, dtype=np.int32)
adata.obs["label"] = y_
#adata.obs["batch"] = b_
batch_key = "batch"
label_key = "label"
adata.uns[batch_key + "_colors"] = [
"#1b9e77",
"#d95f02",
"#7570b3",
"#999999",
"#ff00ff",
] # Set custom colours for batches
#sc.pp.neighbors(adata)
#sc.tl.umap(adata)
#sc.pl.umap(adata, color=[label_key, batch_key], wspace=1, save='batch.pdf')
sc.pp.filter_genes(adata, min_cells=1)
sc.pp.normalize_total(adata)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata)
sc.tl.pca(adata)
sc.pp.neighbors(adata)
sc.tl.louvain(adata)
sc.tl.louvain(adata, key_added = "louvain_1.0") # default resolution in 1.0
true = adata.obs["label"]
best = (0,0,0,0)
for res in np.arange(0.1, 3.01, 0.1):
sc.tl.louvain(adata, resolution = res, key_added = "louvain_"+str(res))
pred = adata.obs['louvain_'+str(res)]
nmi = normalized_mutual_info_score(pred, true, average_method="arithmetic")
label_ari = adjusted_rand_score(pred, y_)
#batch_ari = adjusted_rand_score(pred, b_)
if best[1] < nmi:
best = (res, nmi, -1, label_ari)
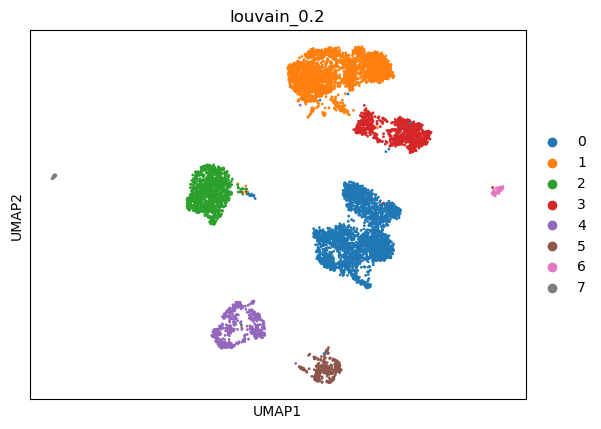
print(best)
(0.2, 0.9181648866263212, -1, 0.9525115188640726)
sc.tl.umap(adata)
sc.pl.umap(adata, color=['louvain_0.2'])
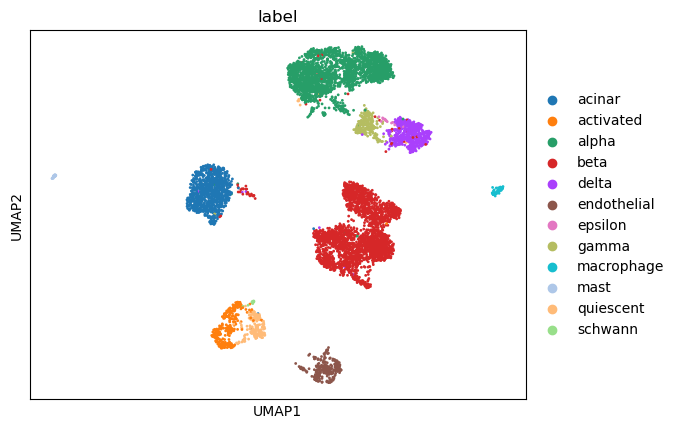
sc.pl.umap(adata, color=['label'])
Muraro: Scanpy-HVG#
datasets = ['./dataset/muraro_sc.h5']
label_filter = ['epsilon', 'alpha', 'beta', 'duct', 'activated', 'schwann', 'gamma', 'quiescent', 'delta', 'macrophage', 'endothelial', 'acinar', 'mast']
X_, y_, b_, file_names = h5_data_loader(datasets, label_filter)
logging.info(f'Data loaded. {datasets}')
adata = ad.AnnData(X_, dtype=np.int32)
adata.obs["label"] = y_
#adata.obs["batch"] = b_
batch_key = "batch"
label_key = "label"
adata.uns[batch_key + "_colors"] = [
"#1b9e77",
"#d95f02",
"#7570b3",
"#999999",
"#ff00ff",
] # Set custom colours for batches
#sc.pp.neighbors(adata)
#sc.tl.umap(adata)
#sc.pl.umap(adata, color=[label_key, batch_key], wspace=1, save='batch.pdf')
sc.pp.filter_genes(adata, min_cells=1)
sc.pp.normalize_total(adata)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata)
sc.tl.pca(adata)
sc.pp.neighbors(adata)
sc.tl.louvain(adata)
sc.tl.louvain(adata, key_added = "louvain_1.0") # default resolution in 1.0
true = adata.obs["label"]
best = (0,0,0,0)
for res in np.arange(0.1, 3.01, 0.1):
sc.tl.louvain(adata, resolution = res, key_added = "louvain_"+str(res))
pred = adata.obs['louvain_'+str(res)]
nmi = normalized_mutual_info_score(pred, true, average_method="arithmetic")
label_ari = adjusted_rand_score(pred, y_)
#batch_ari = adjusted_rand_score(pred, b_)
if best[1] < nmi:
best = (res, nmi, -1, label_ari)
print(best)
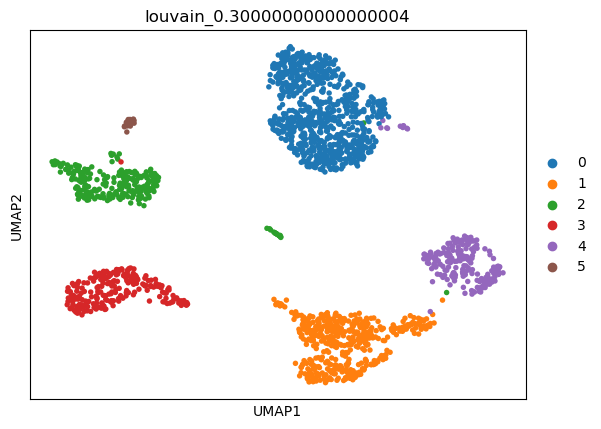
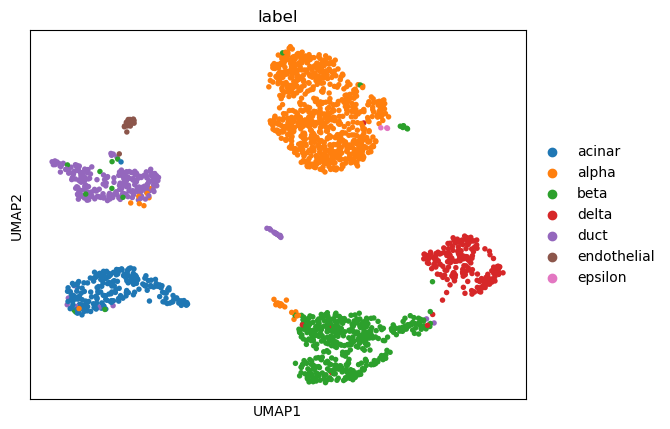
(0.30000000000000004, 0.8825649605389162, -1, 0.9225919369280793)
sc.tl.umap(adata)
sc.pl.umap(adata, color=['louvain_0.30000000000000004'])
sc.pl.umap(adata, color=['label'])
Segerstolpe: Scanpy-HVG#
datasets = ['./dataset/segerstolpe_sc.h5']
label_filter = ['epsilon', 'alpha', 'beta', 'duct', 'activated', 'schwann', 'gamma', 'quiescent', 'delta', 'macrophage', 'endothelial', 'acinar', 'mast']
X_, y_, b_, file_names = h5_data_loader(datasets, label_filter)
logging.info(f'Data loaded. {datasets}')
adata = ad.AnnData(X_, dtype=np.int32)
adata.obs["label"] = y_
#adata.obs["batch"] = b_
batch_key = "batch"
label_key = "label"
adata.uns[batch_key + "_colors"] = [
"#1b9e77",
"#d95f02",
"#7570b3",
"#999999",
"#ff00ff",
] # Set custom colours for batches
#sc.pp.neighbors(adata)
#sc.tl.umap(adata)
#sc.pl.umap(adata, color=[label_key, batch_key], wspace=1, save='batch.pdf')
sc.pp.filter_genes(adata, min_cells=1)
sc.pp.normalize_total(adata)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata)
sc.tl.pca(adata)
sc.pp.neighbors(adata)
sc.tl.louvain(adata)
sc.tl.louvain(adata, key_added = "louvain_1.0") # default resolution in 1.0
true = adata.obs["label"]
best = (0,0,0,0)
for res in np.arange(0.1, 3.01, 0.1):
sc.tl.louvain(adata, resolution = res, key_added = "louvain_"+str(res))
pred = adata.obs['louvain_'+str(res)]
nmi = normalized_mutual_info_score(pred, true, average_method="arithmetic")
label_ari = adjusted_rand_score(pred, y_)
#batch_ari = adjusted_rand_score(pred, b_)
if best[1] < nmi:
best = (res, nmi, -1, label_ari)
print(best)
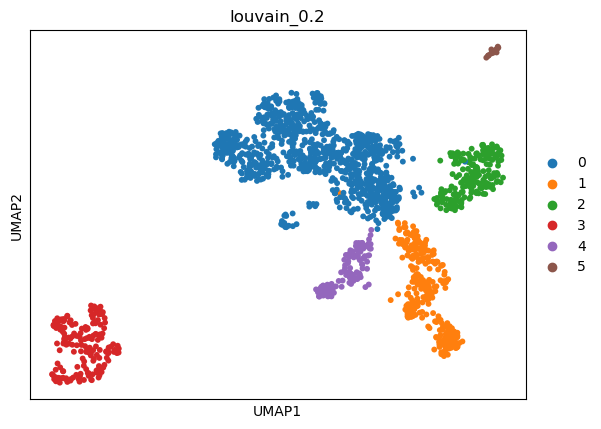
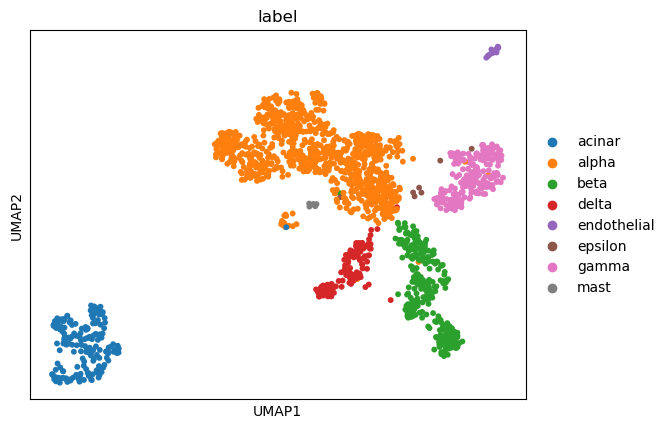
(0.2, 0.9610960547043008, -1, 0.9719308283618725)
sc.tl.umap(adata)
sc.pl.umap(adata, color=['louvain_0.2'])
sc.pl.umap(adata, color=['label'])
Wang: Scanpy-HVG#
datasets = ['./dataset/wang_sc.h5']
label_filter = ['epsilon', 'alpha', 'beta', 'duct', 'activated', 'schwann', 'gamma', 'quiescent', 'delta', 'macrophage', 'endothelial', 'acinar', 'mast']
X_, y_, b_, file_names = h5_data_loader(datasets, label_filter)
logging.info(f'Data loaded. {datasets}')
adata = ad.AnnData(X_, dtype=np.int32)
adata.obs["label"] = y_
#adata.obs["batch"] = b_
batch_key = "batch"
label_key = "label"
adata.uns[batch_key + "_colors"] = [
"#1b9e77",
"#d95f02",
"#7570b3",
"#999999",
"#ff00ff",
] # Set custom colours for batches
#sc.pp.neighbors(adata)
#sc.tl.umap(adata)
#sc.pl.umap(adata, color=[label_key, batch_key], wspace=1, save='batch.pdf')
sc.pp.filter_genes(adata, min_cells=1)
sc.pp.normalize_total(adata)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata)
sc.tl.pca(adata)
sc.pp.neighbors(adata)
sc.tl.louvain(adata)
sc.tl.louvain(adata, key_added = "louvain_1.0") # default resolution in 1.0
true = adata.obs["label"]
best = (0,0,0,0)
for res in np.arange(0.1, 3.01, 0.1):
sc.tl.louvain(adata, resolution = res, key_added = "louvain_"+str(res))
pred = adata.obs['louvain_'+str(res)]
nmi = normalized_mutual_info_score(pred, true, average_method="arithmetic")
label_ari = adjusted_rand_score(pred, y_)
#batch_ari = adjusted_rand_score(pred, b_)
if best[1] < nmi:
best = (res, nmi, -1, label_ari)
print(best)
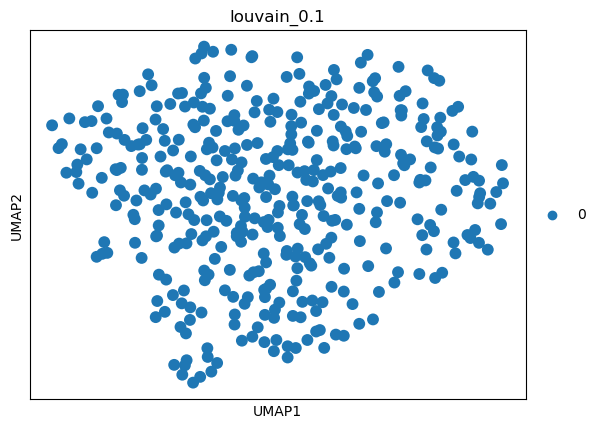
(1.0, 0.5240343182001496, -1, 0.5236983855183179)
sc.tl.umap(adata)
sc.pl.umap(adata, color=['louvain_0.1'])
sc.pl.umap(adata, color=['label'])
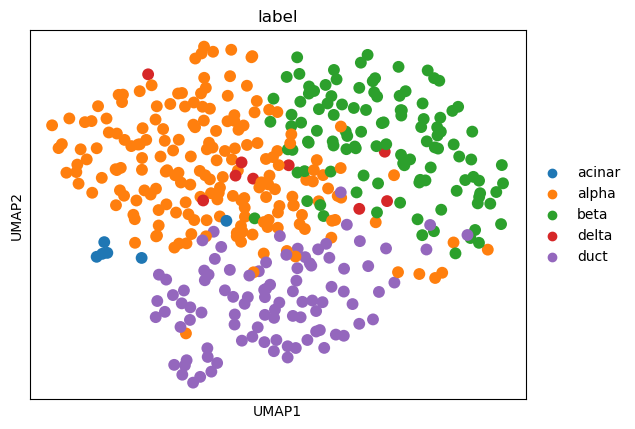
Xin: Scanpy-HVG#
datasets = ['./dataset/xin_sc.h5']
label_filter = ['epsilon', 'alpha', 'beta', 'duct', 'activated', 'schwann', 'gamma', 'quiescent', 'delta', 'macrophage', 'endothelial', 'acinar', 'mast']
X_, y_, b_, file_names = h5_data_loader(datasets, label_filter)
logging.info(f'Data loaded. {datasets}')
adata = ad.AnnData(X_, dtype=np.int32)
adata.obs["label"] = y_
#adata.obs["batch"] = b_
batch_key = "batch"
label_key = "label"
adata.uns[batch_key + "_colors"] = [
"#1b9e77",
"#d95f02",
"#7570b3",
"#999999",
"#ff00ff",
] # Set custom colours for batches
#sc.pp.neighbors(adata)
#sc.tl.umap(adata)
#sc.pl.umap(adata, color=[label_key, batch_key], wspace=1, save='batch.pdf')
sc.pp.filter_genes(adata, min_cells=1)
sc.pp.normalize_total(adata)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata)
sc.tl.pca(adata)
sc.pp.neighbors(adata)
sc.tl.louvain(adata)
sc.tl.louvain(adata, key_added = "louvain_1.0") # default resolution in 1.0
true = adata.obs["label"]
best = (0,0,0,0)
for res in np.arange(0.1, 3.01, 0.1):
sc.tl.louvain(adata, resolution = res, key_added = "louvain_"+str(res))
pred = adata.obs['louvain_'+str(res)]
nmi = normalized_mutual_info_score(pred, true, average_method="arithmetic")
label_ari = adjusted_rand_score(pred, y_)
#batch_ari = adjusted_rand_score(pred, b_)
if best[1] < nmi:
best = (res, nmi, -1, label_ari)
print(best)
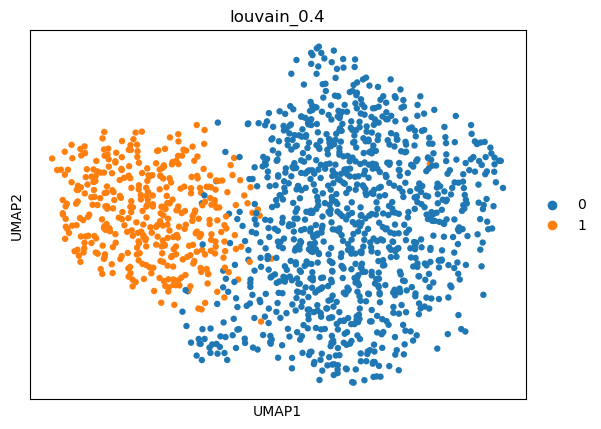
(0.4, 0.626719166892187, -1, 0.6848330417249461)
sc.tl.umap(adata)
sc.pl.umap(adata, color=['louvain_0.4'])
sc.pl.umap(adata, color=['label'])