6-2 Tabular Muris + Baron +MCA (Mouse Pancreas)#
Data load: Tabular_Pancreas + Baron#
import os
import glob
import sys
import pandas as pd
import numpy as np
import random
import matplotlib.pyplot as plt
from sklearn.manifold import TSNE
from umap import UMAP
import seaborn as sns
sys.path.append('../')
import src.utils as my_u
from src.utils import df_cp
from src.utils import df_log
from src.utils import df_total20000
from src.utils import df_minmax
from src.utils import df_l2norm
from src.utils import df_zscore
from src.utils import df_meansquare
from src.utils import run_plot
file_list = glob.glob("../dataset/baron/RAW/*mouse*counts.csv")
print(file_list)
['../dataset/baron/RAW/GSM2230761_mouse1_umifm_counts.csv', '../dataset/baron/RAW/GSM2230762_mouse2_umifm_counts.csv']
total_data = pd.DataFrame()
for ff in file_list:
data = pd.read_csv(ff, sep=',', index_col=0, header=0)
total_data = total_data.append(data)
print(total_data.shape)
labels = total_data['assigned_cluster'].values.tolist()
label_set = set(labels)
print(label_set)
total_data = total_data.iloc[:,2:]
(822, 14880)
(1886, 14880)
{'ductal', 'gamma', 'macrophage', 'immune_other', 'B_cell', 'beta', 'alpha', 'quiescent_stellate', 'activated_stellate', 'T_cell', 'schwann', 'endothelial', 'delta'}
baron_labels = labels
baron_total_data = total_data
Data load: Tabula Pancreas#
200 cells are sampled
file_list = glob.glob("../data/tabular_muris/00_facs_raw_data/FACS/Pancreas-counts.csv")
sample_per_class = 200
labels = []
total_data = pd.DataFrame()
for ff in file_list:
data = pd.read_csv(ff, sep=',', index_col=0, header=0)
data = data.transpose()
c = list(range(data.shape[0]))
total_data = pd.concat([total_data, data.iloc[c,]], axis=0)
labels += [ff.split('/')[-1].rstrip('-counts.csv')] * sample_per_class #data.shape[0]
label_set = set(labels)
print(label_set)
annot_label = pd.read_csv('../data/tabular_muris/00_facs_raw_data/annotations_FACS.csv', sep=',', index_col=0, header=0)
com = annot_label['tissue'].index.intersection(total_data.index)
m = annot_label.filter(com, axis=0)
total_data = total_data.filter(com, axis=0)
total_data = pd.concat([total_data, m['cell_ontology_class']],axis =1)
tissue = labels
labels = total_data['cell_ontology_class']
labels = labels.values.tolist()
total_data.pop('cell_ontology_class')
total_data.pop('zsGreen_transgene')
print(set(labels))
{'Pancrea'}
{'pancreatic PP cell', 'pancreatic stellate cell', 'leukocyte', 'endothelial cell', 'pancreatic D cell', 'pancreatic ductal cell', 'pancreatic acinar cell', 'type B pancreatic cell', 'pancreatic A cell'}
tabula_data = total_data
tabula_labels = labels
new_lab = []
for x in tabula_labels:
try:
x=x.replace('pancreatic A cell', 'alpha')
except:
pass
try:
x=x.replace('endothelial cell', 'endothelial')
except:
pass
try:
x=x.replace('pancreatic D cell', 'delta')
except:
pass
try:
x=x.replace('pancreatic acinar cell', 'acinar')
except:
pass
try:
x=x.replace('pancreatic ductal cell', 'ductal')
except:
pass
try:
x=x.replace('type B pancreatic cell', 'beta')
except:
pass
try:
x=x.replace('pancreatic stellate cell', 'stellate')
except:
pass
try:
x=x.replace('pancreatic PP cell', 'PP')
except:
pass
try:
x=x.replace('activated_stellate', 'stellate')
except:
pass
try:
x=x.replace('quiescent_stellate', 'stellate')
except:
pass
new_lab.append(x)
tabula_labels = new_lab
print(set(tabula_labels))
{'ductal', 'leukocyte', 'beta', 'alpha', 'stellate', 'acinar', 'endothelial', 'PP', 'delta'}
Data load: MCA#
common_mca_files = glob.glob("../dataset/MCA/mousecellatlas/*_Pancreas_*.txt")
print((common_mca_files))
['../dataset/MCA/mousecellatlas/GSM2906458_Pancreas_dge.txt']
# MCA data load
#sample_per_class = 500
mca_labels = []
mca_tabula_labels = []
mca_data = pd.DataFrame()
for ff in common_mca_files:
data = pd.read_csv(ff, sep='\s+', index_col=0, header=0)
data = data.transpose()
c = list(range(data.shape[0]))
data = data.iloc[c,]
mca_data = pd.concat([mca_data, data], axis=0)
print(mca_data.shape, ff)
mca_labels += [ff.rstrip('_dge.txt').split('/')[-1].split('_')[1]] * data.shape[0]
mca_data.replace(np.nan,0)
mca_label_set = set(mca_labels)
print(mca_label_set)
(4405, 20283) ../dataset/MCA/mousecellatlas/GSM2906458_Pancreas_dge.txt
{'Pancreas'}
m = pd.read_csv("../dataset/MCA/mousecellatlas/MCA_CellAssignments.csv", sep=',', index_col=1, header=0)
#print(m)
#m = m.replace(np.nan, 'nan')
#m = m.loc[m['Annotation'] != 'nan',]
print(m.shape)
m = m.loc[m['Annotation'] != 'Dividing cell(Pancreas)',]
m = m.loc[m['Annotation'] != 'Erythroblast_Igkc high(Pancreas)',]
m = m.loc[m['Annotation'] != 'Erythroblast_Hbb-bt high(Pancreas)',]
m = m.loc[m['Annotation'] != 'Stromal cell_Fn1 high(Pancreas)',]
m = m.loc[m['Annotation'] != 'Stromal cell_Smoc2 high(Pancreas)',]
m = m.loc[m['Annotation'] != 'Stromal cell_Mfap4 high(Pancreas)',]
m = m.loc[m['Annotation'] != 'Smooth muscle cell_Acta2 high(Pancreas)',]
m = m.loc[m['Annotation'] != 'Smooth muscle cell_Rgs5 high(Pancreas)',]
print(m.shape)
com = m['Annotation'].index.intersection(mca_data.index)
m = m.filter(com, axis=0)
mca_data = mca_data.filter(com, axis=0)
print(m.shape)
mca_data = pd.concat([mca_data, m[['Tissue','Annotation']]],axis =1)
mca_tissue = mca_data['Tissue']
mca_labels = mca_data['Annotation']
mca_tissue = mca_tissue.values.tolist()
mca_labels = mca_labels.values.tolist()
mca_data.pop('Tissue')
mca_data.pop('Annotation')
print(set(list(mca_labels)))
print(set(mca_tissue))
(270848, 6)
(269196, 6)
(1958, 6)
{'β-cell(Pancreas)', 'Ductal cell(Pancreas)', 'Endothelial cell_Fabp4 high(Pancreas)', 'Macrophage_Ly6c2 high(Pancreas)', 'Glial cell(Pancreas)', 'Macrophage(Pancreas)', 'Endocrine cell(Pancreas)', 'B cell(Pancreas)', 'Acinar cell(Pancreas)', 'Endothelial cell_Lrg1 high(Pancreas)', 'T cell(Pancreas)', 'Endothelial cell_Tm4sf1 high(Pancreas)', 'Granulocyte(Pancreas)', 'Dendrtic cell(Pancreas)'}
{'Pancreas'}
mca_labels = [x.split('_')[0].split('(')[0] for x in mca_labels]
mca_data.shape
print(set(mca_labels))
{'Granulocyte', 'β-cell', 'Acinar cell', 'Ductal cell', 'B cell', 'Endocrine cell', 'Endothelial cell', 'Dendrtic cell', 'T cell', 'Macrophage', 'Glial cell'}
common_gene = baron_total_data.columns.intersection(tabula_data.columns)
common_gene = common_gene.intersection(mca_data.columns)
tabula_data = tabula_data.filter(common_gene)
baron_total_data = baron_total_data.filter(common_gene)
mca_data = mca_data.filter(common_gene)
total_data = pd.concat([tabula_data,baron_total_data, mca_data], axis=0)
labels = tabula_labels + baron_labels + mca_labels
blabels = ['tabula']*len(tabula_labels) + ['baron']*len(baron_labels) + ['MCA'] * len(mca_labels)
total_data.shape
print(total_data.shape)
print(set(labels))
print(set(blabels))
(5171, 12584)
{'Granulocyte', 'B cell', 'Endothelial cell', 'leukocyte', 'alpha', 'activated_stellate', 'gamma', 'ductal', 'Acinar cell', 'B_cell', 'T cell', 'Glial cell', 'T_cell', 'acinar', 'Ductal cell', 'Endocrine cell', 'immune_other', 'Dendrtic cell', 'beta', 'stellate', 'schwann', 'endothelial', 'delta', 'β-cell', 'macrophage', 'quiescent_stellate', 'Macrophage', 'PP'}
{'MCA', 'tabula', 'baron'}
new_lab2 = []
for x in labels:
try:
x=x.replace('Endothelial cell', 'endothelial')
except:
continue
try:
x=x.replace('Glial cell', 'i_Glial')
except:
continue
try:
x=x.replace('Granulocyte', 'i_Granulocyte')
except:
continue
try:
x=x.replace('Dendrtic cell', 'i_dendrtic_cell')
except:
continue
try:
x=x.replace('Ductal cell', 'ductal')
except:
continue
try:
x=x.replace('Acinar cell', 'acinar')
except:
continue
try:
x=x.replace('β-cell', 'beta')
except:
continue
try:
x=x.replace('B cell', 'i_b_cell')
except:
continue
try:
x=x.replace('B_cell', 'i_b_cell')
except:
continue
try:
x=x.replace('T cell', 'i_t_cell')
except:
continue
try:
x=x.replace('T_cell', 'i_t_cell')
except:
continue
try:
x=x.replace('gamma', 'i_gamma')
except:
continue
try:
x=x.replace('Myeloid progenitor cell', 'i_Myeloid progenitor cell')
except:
continue
try:
x=x.replace('Monocyte', 'i_Monocyte')
except:
continue
try:
x=x.replace('NK cell', 'i_NK_cell')
except:
continue
try:
x=x.replace('leukocyte', 'i_leukocyte')
except:
continue
try:
x=x.replace('macrophage', 'i_macrophage')
except:
continue
try:
x=x.replace('Macrophage', 'i_macrophage')
except:
continue
try:
x=x.replace('Dendritic cell', 'i_Dendritic cell')
except:
continue
try:
x=x.replace('activated_stellate', 'stellate')
except:
continue
try:
x=x.replace('quiescent_stellate', 'stellate')
except:
continue
try:
x=x.replace('pancreatic PP cell', 'e_PP_cell')
except:
continue
new_lab2.append(x)
labels2 = new_lab2
print(set(labels2))
{'i_macrophage', 'alpha', 'i_Granulocyte', 'ductal', 'i_leukocyte', 'i_dendrtic_cell', 'acinar', 'i_gamma', 'i_t_cell', 'Endocrine cell', 'immune_other', 'beta', 'stellate', 'i_Glial', 'schwann', 'endothelial', 'delta', 'i_b_cell', 'PP'}
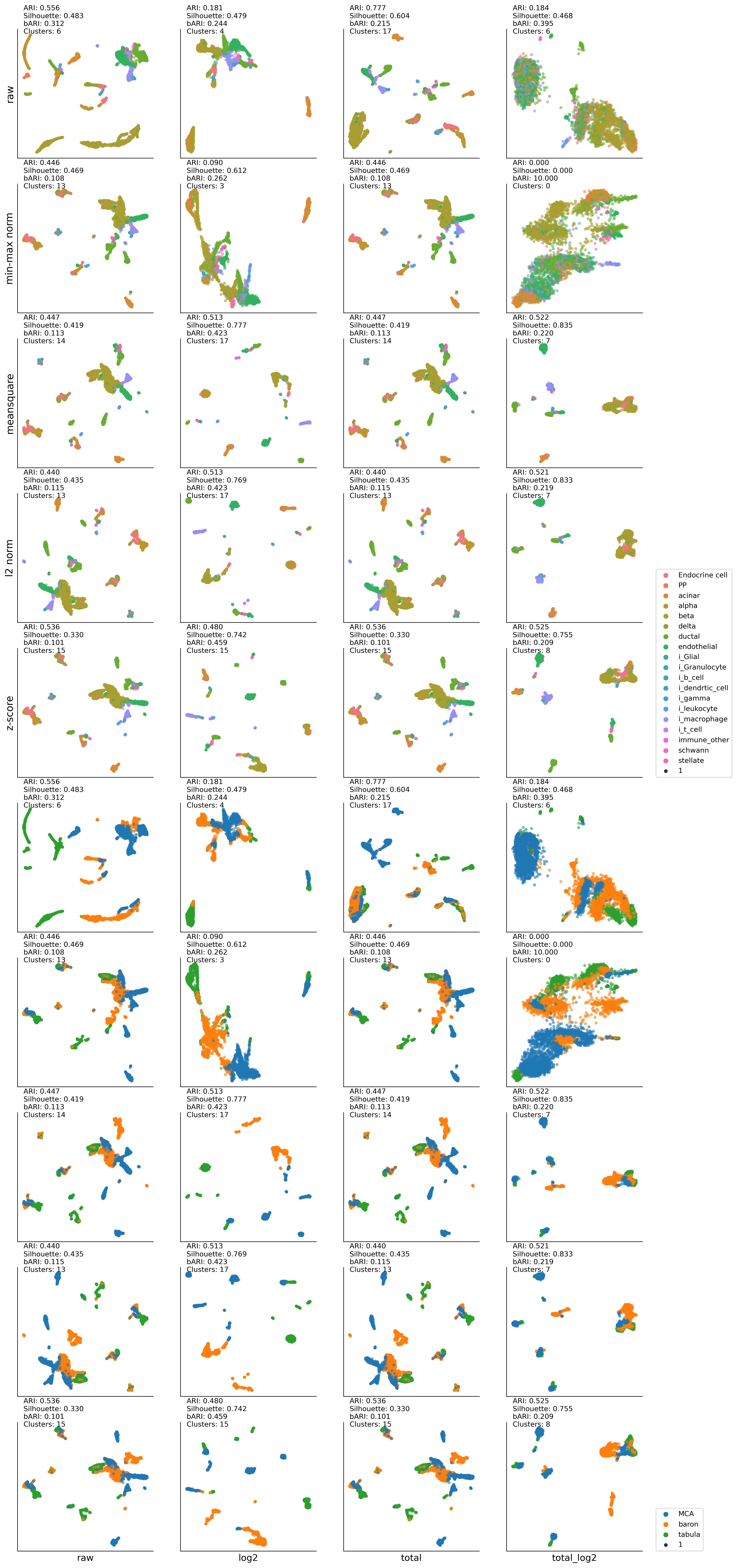
t-SNE:DBSCAN results#
latent_space = TSNE(n_components=2)
#latent_space = UMAP(n_components=2, init='spectral', random_state=0)
clustering_method = 'dbscan'
############################################
plt.figure(figsize=(16,40), dpi=300)
ax00 = plt.subplot2grid((10,4), (0,0))
ax10 = plt.subplot2grid((10,4), (0,1))
ax20 = plt.subplot2grid((10,4), (0,2))
ax30 = plt.subplot2grid((10,4), (0,3))
ax01 = plt.subplot2grid((10,4), (1,0))
ax11 = plt.subplot2grid((10,4), (1,1))
ax21 = plt.subplot2grid((10,4), (1,2))
ax31 = plt.subplot2grid((10,4), (1,3))
ax02 = plt.subplot2grid((10,4), (2,0))
ax12 = plt.subplot2grid((10,4), (2,1))
ax22 = plt.subplot2grid((10,4), (2,2))
ax32 = plt.subplot2grid((10,4), (2,3))
ax03 = plt.subplot2grid((10,4), (3,0))
ax13 = plt.subplot2grid((10,4), (3,1))
ax23 = plt.subplot2grid((10,4), (3,2))
ax33 = plt.subplot2grid((10,4), (3,3))
ax04 = plt.subplot2grid((10,4), (4,0))
ax14 = plt.subplot2grid((10,4), (4,1))
ax24 = plt.subplot2grid((10,4), (4,2))
ax34 = plt.subplot2grid((10,4), (4,3))
ax05 = plt.subplot2grid((10,4), (5,0))
ax15 = plt.subplot2grid((10,4), (5,1))
ax25 = plt.subplot2grid((10,4), (5,2))
ax35 = plt.subplot2grid((10,4), (5,3))
ax06 = plt.subplot2grid((10,4), (6,0))
ax16 = plt.subplot2grid((10,4), (6,1))
ax26 = plt.subplot2grid((10,4), (6,2))
ax36 = plt.subplot2grid((10,4), (6,3))
ax07 = plt.subplot2grid((10,4), (7,0))
ax17 = plt.subplot2grid((10,4), (7,1))
ax27 = plt.subplot2grid((10,4), (7,2))
ax37 = plt.subplot2grid((10,4), (7,3))
ax08 = plt.subplot2grid((10,4), (8,0))
ax18 = plt.subplot2grid((10,4), (8,1))
ax28 = plt.subplot2grid((10,4), (8,2))
ax38 = plt.subplot2grid((10,4), (8,3))
ax09 = plt.subplot2grid((10,4), (9,0))
ax19 = plt.subplot2grid((10,4), (9,1))
ax29 = plt.subplot2grid((10,4), (9,2))
ax39 = plt.subplot2grid((10,4), (9,3))
l = []
############################################
latent_space = TSNE(n_components=2)
l.append(run_plot(df_cp(total_data), \
ax00, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax05)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_log(df_cp(total_data)), \
ax10, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax15)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_total20000(df_cp(total_data)), \
ax20, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax25)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_log(df_total20000(df_cp(total_data))), \
ax30, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax35)
)
############################################
latent_space = TSNE(n_components=2)
l.append(run_plot(df_minmax(df_cp(total_data)), \
ax01, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax06)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_minmax(df_log(df_cp(total_data))), \
ax11, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax16)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_minmax(df_total20000(df_cp(total_data))), \
ax21, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax26)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_minmax(df_log(df_total20000(df_cp(total_data)))), \
ax31, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax36)
)
############################################
latent_space = TSNE(n_components=2)
l.append(run_plot(df_meansquare(df_cp(total_data)), \
ax02, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax07)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_meansquare(df_log(df_cp(total_data))), \
ax12, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax17)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_meansquare(df_total20000(df_cp(total_data))), \
ax22, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax27)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_meansquare(df_log(df_total20000(df_cp(total_data)))), \
ax32, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax37)
)
############################################
latent_space = TSNE(n_components=2)
l.append(run_plot(df_l2norm(df_cp(total_data)), \
ax03, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax08)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_l2norm(df_log(df_cp(total_data))), \
ax13, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax18)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_l2norm(df_total20000(df_cp(total_data))), \
ax23, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax28)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_l2norm(df_log(df_total20000(df_cp(total_data)))), \
ax33, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax38)
)
############################################
latent_space = TSNE(n_components=2)
l.append(run_plot(df_zscore(df_cp(total_data)), \
ax04, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax09)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_zscore(df_log(df_cp(total_data))), \
ax14, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax19)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_zscore(df_total20000(df_cp(total_data))), \
ax24, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax29)
)
latent_space = TSNE(n_components=2)
l.append(run_plot(df_zscore(df_log(df_total20000(df_cp(total_data)))), \
ax34, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax39)
)
############################################
ax00.set_ylabel('raw' , fontsize=14)
ax01.set_ylabel('min-max norm' , fontsize=14)
ax02.set_ylabel('meansquare' , fontsize=14)
ax03.set_ylabel('l2 norm' , fontsize=14)
ax04.set_ylabel('z-score' , fontsize=14)
ax09.set_xlabel('raw', fontsize=13)
ax19.set_xlabel('log2', fontsize=13)
ax29.set_xlabel('total', fontsize=13)
ax39.set_xlabel('total_log2', fontsize=13)
ax30.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
ax39.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
dbscan #cluster: (24, 0.5591372870952797, 0.14259721, 0.3163442353929353)
dbscan #cluster: (24, 0.3444734032569326, 0.0037780951, 0.19505786975681375)
dbscan #cluster: (21, 0.775519963482579, 0.28476873, 0.20771075925684437)
dbscan #cluster: (25, 0.2219069412344603, -0.17520925, 0.20774876222517227)
dbscan #cluster: (31, 0.6349873931882888, 0.2570556, 0.20857468948073948)
dbscan #cluster: (18, 0.2981544362651802, 0.10989848, 0.2718158595307432)
dbscan #cluster: (26, 0.6189605734552697, 0.2815414, 0.2073348437861064)
dbscan #cluster: (20, 0.18137684727112552, -0.14150067, 0.12655689141839552)
dbscan #cluster: (29, 0.6441315700319818, 0.27123496, 0.21749977192778133)
dbscan #cluster: (22, 0.48545146991182503, 0.54844433, 0.36161006931808487)
dbscan #cluster: (30, 0.6400837540616197, 0.291539, 0.21263431736930974)
dbscan #cluster: (7, 0.5029349219599313, 0.52430564, 0.1696391683415515)
dbscan #cluster: (32, 0.6404563961801408, 0.26591963, 0.21296838957743533)
dbscan #cluster: (23, 0.4885719797916577, 0.5550364, 0.3636765519580801)
dbscan #cluster: (31, 0.6368464456860983, 0.2280137, 0.2142755309512389)
dbscan #cluster: (7, 0.5136385753653934, 0.559173, 0.17547125793643684)
dbscan #cluster: (31, 0.6373788059572258, 0.2862702, 0.2139018550109023)
dbscan #cluster: (28, 0.5050321990335069, 0.5839211, 0.29822095652555936)
dbscan #cluster: (30, 0.6491363894037523, 0.26433417, 0.21846438454431533)
dbscan #cluster: (14, 0.4574689736882003, 0.42957127, 0.1607634528520651)
<matplotlib.legend.Legend at 0x7f661c3578e0>
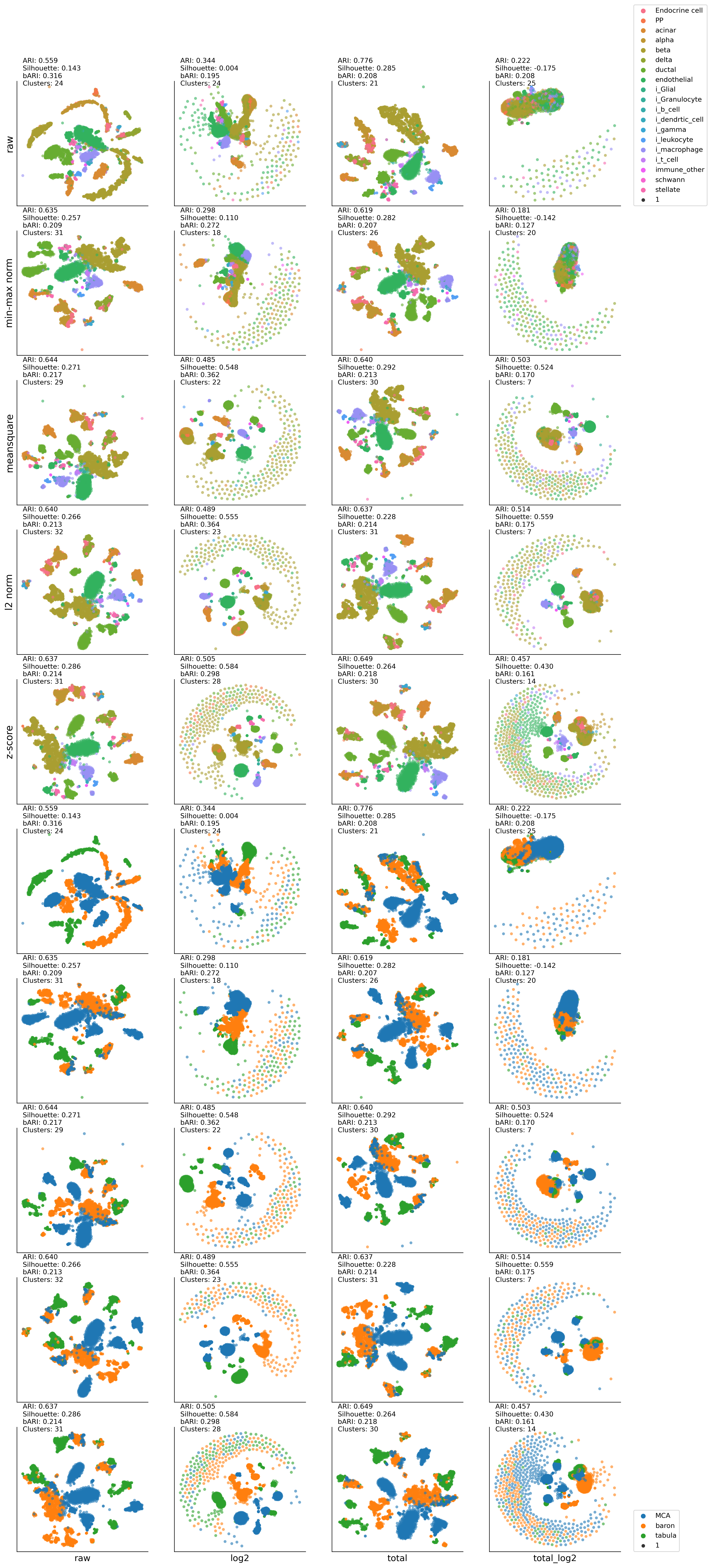
UMAP:DBSCAN results#
clustering_method = 'dbscan'
############################################
plt.figure(figsize=(16,40), dpi=300)
ax00 = plt.subplot2grid((10,4), (0,0))
ax10 = plt.subplot2grid((10,4), (0,1))
ax20 = plt.subplot2grid((10,4), (0,2))
ax30 = plt.subplot2grid((10,4), (0,3))
ax01 = plt.subplot2grid((10,4), (1,0))
ax11 = plt.subplot2grid((10,4), (1,1))
ax21 = plt.subplot2grid((10,4), (1,2))
ax31 = plt.subplot2grid((10,4), (1,3))
ax02 = plt.subplot2grid((10,4), (2,0))
ax12 = plt.subplot2grid((10,4), (2,1))
ax22 = plt.subplot2grid((10,4), (2,2))
ax32 = plt.subplot2grid((10,4), (2,3))
ax03 = plt.subplot2grid((10,4), (3,0))
ax13 = plt.subplot2grid((10,4), (3,1))
ax23 = plt.subplot2grid((10,4), (3,2))
ax33 = plt.subplot2grid((10,4), (3,3))
ax04 = plt.subplot2grid((10,4), (4,0))
ax14 = plt.subplot2grid((10,4), (4,1))
ax24 = plt.subplot2grid((10,4), (4,2))
ax34 = plt.subplot2grid((10,4), (4,3))
ax05 = plt.subplot2grid((10,4), (5,0))
ax15 = plt.subplot2grid((10,4), (5,1))
ax25 = plt.subplot2grid((10,4), (5,2))
ax35 = plt.subplot2grid((10,4), (5,3))
ax06 = plt.subplot2grid((10,4), (6,0))
ax16 = plt.subplot2grid((10,4), (6,1))
ax26 = plt.subplot2grid((10,4), (6,2))
ax36 = plt.subplot2grid((10,4), (6,3))
ax07 = plt.subplot2grid((10,4), (7,0))
ax17 = plt.subplot2grid((10,4), (7,1))
ax27 = plt.subplot2grid((10,4), (7,2))
ax37 = plt.subplot2grid((10,4), (7,3))
ax08 = plt.subplot2grid((10,4), (8,0))
ax18 = plt.subplot2grid((10,4), (8,1))
ax28 = plt.subplot2grid((10,4), (8,2))
ax38 = plt.subplot2grid((10,4), (8,3))
ax09 = plt.subplot2grid((10,4), (9,0))
ax19 = plt.subplot2grid((10,4), (9,1))
ax29 = plt.subplot2grid((10,4), (9,2))
ax39 = plt.subplot2grid((10,4), (9,3))
l = []
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_cp(total_data), \
ax00, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax05)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_cp(total_data)), \
ax10, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax15)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_total20000(df_cp(total_data)), \
ax20, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax25)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_log(df_total20000(df_cp(total_data))), \
ax30, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax35)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_cp(total_data)), \
ax01, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax06)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_cp(total_data))), \
ax11, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax16)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_total20000(df_cp(total_data))), \
ax21, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax26)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_minmax(df_log(df_total20000(df_cp(total_data)))), \
ax31, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax36)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_cp(total_data)), \
ax02, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax07)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_cp(total_data))), \
ax12, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax17)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_total20000(df_cp(total_data))), \
ax22, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax27)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_meansquare(df_log(df_total20000(df_cp(total_data)))), \
ax32, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax37)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_cp(total_data)), \
ax03, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax08)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_cp(total_data))), \
ax13, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax18)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_total20000(df_cp(total_data))), \
ax23, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax28)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_l2norm(df_log(df_total20000(df_cp(total_data)))), \
ax33, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax38)
)
############################################
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_cp(total_data)), \
ax04, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax09)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_cp(total_data))), \
ax14, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax19)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_total20000(df_cp(total_data))), \
ax24, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax29)
)
latent_space = UMAP(n_components=2, init='spectral', random_state=0)
l.append(run_plot(df_zscore(df_log(df_total20000(df_cp(total_data)))), \
ax34, labels2, latent_space, clustering_method, blabels=blabels, b_ax=ax39)
)
############################################
ax00.set_ylabel('raw' , fontsize=14)
ax01.set_ylabel('min-max norm' , fontsize=14)
ax02.set_ylabel('meansquare' , fontsize=14)
ax03.set_ylabel('l2 norm' , fontsize=14)
ax04.set_ylabel('z-score' , fontsize=14)
ax09.set_xlabel('raw', fontsize=13)
ax19.set_xlabel('log2', fontsize=13)
ax29.set_xlabel('total', fontsize=13)
ax39.set_xlabel('total_log2', fontsize=13)
ax34.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
ax39.legend(bbox_to_anchor=(1.1,0), loc='lower left',borderaxespad=0)
dbscan #cluster: (6, 0.5559543348730758, 0.48291337, 0.31190072241376765)
dbscan #cluster: (4, 0.1814924933510689, 0.47886303, 0.24438230822947316)
dbscan #cluster: (17, 0.7765156117748234, 0.6039262, 0.21494128733605739)
dbscan #cluster: (6, 0.18360780761235834, 0.46764058, 0.39469556372136133)
dbscan #cluster: (13, 0.4458622368537242, 0.4685685, 0.10843824139226949)
dbscan #cluster: (3, 0.08990909443428714, 0.61157554, 0.2621915912791283)
dbscan #cluster: (13, 0.4458622368537242, 0.4685685, 0.10843824139226949)
dbscan #cluster: (0, 0, 0, 10)
dbscan #cluster: (14, 0.4470966008898146, 0.41907063, 0.11320547077325722)
dbscan #cluster: (17, 0.5129220243077792, 0.77690804, 0.42277184647695576)
dbscan #cluster: (14, 0.4470966008898146, 0.41907063, 0.11320547077325722)
dbscan #cluster: (7, 0.5215438153646637, 0.8351135, 0.21980901127846342)
dbscan #cluster: (13, 0.44014421169874235, 0.4345569, 0.11504732193594494)
dbscan #cluster: (17, 0.5128181407449381, 0.76890224, 0.42325396196949383)
dbscan #cluster: (13, 0.44014421169874235, 0.4345569, 0.11504732193594494)
dbscan #cluster: (7, 0.5214205353597122, 0.8334219, 0.21896889478988615)
dbscan #cluster: (15, 0.5361025602416168, 0.3299337, 0.10124821472556857)
dbscan #cluster: (15, 0.47959156079504195, 0.7417183, 0.45907280442307974)
dbscan #cluster: (15, 0.5361025602416168, 0.3299337, 0.10124821472556857)
dbscan #cluster: (8, 0.5248193999905937, 0.75469875, 0.20861942517469187)
<matplotlib.legend.Legend at 0x7f6617e687c0>