5. Pancreas dataset integration with scVI/scANVI/BBKNN#
Low-dimensional integration will be compared with scVI and scANVI
Referenced pages.
https://www.sc-best-practices.org/preamble.html
https://satijalab.org/seurat/articles/integration_mapping
!pip install --quiet scanpy
!pip install --quiet 'scanpy[leiden]'
!pip install --quiet louvain
!pip install --quiet scvi-colab
from scvi_colab import install
install()
import warnings
warnings.filterwarnings('ignore')
import os
import tempfile
import sys
sys.path.append('/content/drive/MyDrive/Colab Notebooks/')
sys.path.append('/content/drive/MyDrive/Colab Notebooks/src')
import utils as my_u
from utils import data_transformation
from utils import clustering
from utils import h5_data_loader
import anndata as ad
import scanpy as sc
import logging
import numpy as np
from sklearn.decomposition import PCA
from sklearn import preprocessing
from sklearn.metrics.cluster import normalized_mutual_info_score
from sklearn.metrics.cluster import adjusted_rand_score
import umap
import scvi
import torch
sys.path.append('/content/drive/MyDrive/Colab Notebooks/')
datasets = ['/content/drive/MyDrive/Colab Notebooks/dataset/baron_sc.h5', '/content/drive/MyDrive/Colab Notebooks/dataset/muraro_sc.h5', '/content/drive/MyDrive/Colab Notebooks/dataset/segerstolpe_sc.h5', '/content/drive/MyDrive/Colab Notebooks/dataset/wang_sc.h5', '/content/drive/MyDrive/Colab Notebooks/dataset/xin_sc.h5']
label_filter = ['epsilon', 'alpha', 'beta', 'duct', 'activated', 'schwann', 'gamma', 'quiescent', 'delta', 'macrophage', 'endothelial', 'acinar', 'mast']
scVI with Total->Log preprocessing#
scVI integration work
X_, y_, b_, file_names = h5_data_loader(datasets, label_filter)
logging.info(f'Data loaded. {datasets}')
adata = ad.AnnData(X_, dtype=np.int32)
adata.obs["label"] = y_
adata.obs["batch"] = b_
batch_key = "batch"
label_key = "label"
adata.uns[batch_key + "_colors"] = [
"#1b9e77",
"#d95f02",
"#7570b3",
"#999999",
"#ff00ff",
] # Set custom colours for batches
sc.pp.filter_genes(adata, min_cells=2)
adata.layers["counts"] = adata.X.copy()
sc.pp.normalize_total(adata)
sc.pp.log1p(adata)
adata.layers["logcounts"] = adata.X.copy()
sc.tl.pca(adata)
#sc.pp.neighbors(adata)
adata_scvi = adata.copy()
scvi.model.SCVI.setup_anndata(adata_scvi, layer="counts", batch_key=batch_key)
model_scvi = scvi.model.SCVI(adata_scvi)
model_scvi.view_anndata_setup()
Anndata setup with scvi-tools version 1.0.3.
Setup via `SCVI.setup_anndata` with arguments:
{ │ 'layer': 'counts', │ 'batch_key': 'batch', │ 'labels_key': None, │ 'size_factor_key': None, │ 'categorical_covariate_keys': None, │ 'continuous_covariate_keys': None }
Summary Statistics ┏━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━┓ ┃ Summary Stat Key ┃ Value ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━┩ │ n_batch │ 5 │ │ n_cells │ 12927 │ │ n_extra_categorical_covs │ 0 │ │ n_extra_continuous_covs │ 0 │ │ n_labels │ 1 │ │ n_vars │ 15590 │ └──────────────────────────┴───────┘
Data Registry ┏━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Registry Key ┃ scvi-tools Location ┃ ┡━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ X │ adata.layers['counts'] │ │ batch │ adata.obs['_scvi_batch'] │ │ labels │ adata.obs['_scvi_labels'] │ └──────────────┴───────────────────────────┘
batch State Registry ┏━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['batch'] │ baron_sc │ 0 │ │ │ muraro_sc │ 1 │ │ │ segerstolpe_sc │ 2 │ │ │ wang_sc │ 3 │ │ │ xin_sc │ 4 │ └────────────────────┴────────────────┴─────────────────────┘
labels State Registry ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['_scvi_labels'] │ 0 │ 0 │ └───────────────────────────┴────────────┴─────────────────────┘
max_epochs_scvi = np.min([round((20000 / adata.n_obs) * 400), 400])
model_scvi.train()
INFO: GPU available: True (cuda), used: True
INFO:lightning.pytorch.utilities.rank_zero:GPU available: True (cuda), used: True
INFO: TPU available: False, using: 0 TPU cores
INFO:lightning.pytorch.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO: IPU available: False, using: 0 IPUs
INFO:lightning.pytorch.utilities.rank_zero:IPU available: False, using: 0 IPUs
INFO: HPU available: False, using: 0 HPUs
INFO:lightning.pytorch.utilities.rank_zero:HPU available: False, using: 0 HPUs
INFO: LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
INFO:lightning.pytorch.accelerators.cuda:LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 400/400: 100%|██████████| 400/400 [10:46<00:00, 1.51s/it, v_num=1, train_loss_step=1.31e+4, train_loss_epoch=1.19e+4]
INFO: `Trainer.fit` stopped: `max_epochs=400` reached.
INFO:lightning.pytorch.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=400` reached.
Epoch 400/400: 100%|██████████| 400/400 [10:46<00:00, 1.62s/it, v_num=1, train_loss_step=1.31e+4, train_loss_epoch=1.19e+4]
adata_scvi.obsm["X_scVI"] = model_scvi.get_latent_representation()
sc.pp.neighbors(adata_scvi, use_rep="X_scVI")
sc.tl.umap(adata_scvi)
adata_scvi
sc.pl.umap(adata_scvi, color=[label_key, batch_key], wspace=1)

sc.pp.neighbors(adata_scvi, use_rep='X_scVI', key_added = "scvi_nn")
true = adata.obs["label"]
best = (0,0,0,0)
for res in np.arange(0.1, 3.01, 0.1):
sc.tl.louvain(adata_scvi, obsp='scvi_nn_distances', resolution = res, key_added = "louvain_scvi_"+str(res))
pred = adata_scvi.obs['louvain_scvi_'+str(res)]
nmi = normalized_mutual_info_score(pred, true, average_method="arithmetic")
label_ari = adjusted_rand_score(pred, y_)
batch_ari = adjusted_rand_score(pred, b_)
if best[1] < nmi:
best = (res, nmi, batch_ari, label_ari)
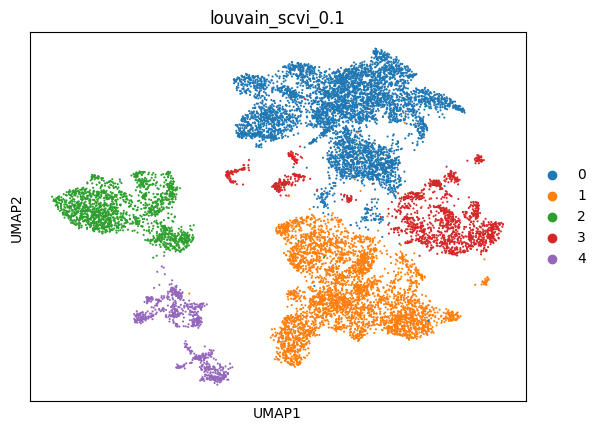
print(best)
(0.1, 0.8484036108793291, -0.01931710040087526, 0.9204421528807389)
sc.tl.umap(adata_scvi)
sc.pl.umap(adata_scvi, color=['louvain_scvi_0.1'])
scVI with Total preprocessing#
X_, y_, b_, file_names = h5_data_loader(datasets, label_filter)
logging.info(f'Data loaded. {datasets}')
adata = ad.AnnData(X_, dtype=np.int32)
adata.obs["label"] = y_
adata.obs["batch"] = b_
batch_key = "batch"
label_key = "label"
adata.uns[batch_key + "_colors"] = [
"#1b9e77",
"#d95f02",
"#7570b3",
"#999999",
"#ff00ff",
] # Set custom colours for batches
sc.pp.filter_genes(adata, min_cells=2)
adata.layers["counts"] = adata.X.copy()
sc.pp.normalize_total(adata)
#sc.pp.log1p(adata)
adata.layers["logcounts"] = adata.X.copy()
sc.tl.pca(adata)
#sc.pp.neighbors(adata)
adata_scvi = adata.copy()
scvi.model.SCVI.setup_anndata(adata_scvi, layer="counts", batch_key=batch_key)
model_scvi = scvi.model.SCVI(adata_scvi)
model_scvi.view_anndata_setup()
max_epochs_scvi = np.min([round((20000 / adata.n_obs) * 400), 400])
model_scvi.train()
adata_scvi.obsm["X_scVI"] = model_scvi.get_latent_representation()
sc.pp.neighbors(adata_scvi, use_rep="X_scVI")
sc.tl.umap(adata_scvi)
adata_scvi
sc.pl.umap(adata_scvi, color=[label_key, batch_key], wspace=1)
Anndata setup with scvi-tools version 1.0.3.
Setup via `SCVI.setup_anndata` with arguments:
{ │ 'layer': 'counts', │ 'batch_key': 'batch', │ 'labels_key': None, │ 'size_factor_key': None, │ 'categorical_covariate_keys': None, │ 'continuous_covariate_keys': None }
Summary Statistics ┏━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━┓ ┃ Summary Stat Key ┃ Value ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━┩ │ n_batch │ 5 │ │ n_cells │ 12927 │ │ n_extra_categorical_covs │ 0 │ │ n_extra_continuous_covs │ 0 │ │ n_labels │ 1 │ │ n_vars │ 15590 │ └──────────────────────────┴───────┘
Data Registry ┏━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Registry Key ┃ scvi-tools Location ┃ ┡━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ X │ adata.layers['counts'] │ │ batch │ adata.obs['_scvi_batch'] │ │ labels │ adata.obs['_scvi_labels'] │ └──────────────┴───────────────────────────┘
batch State Registry ┏━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['batch'] │ baron_sc │ 0 │ │ │ muraro_sc │ 1 │ │ │ segerstolpe_sc │ 2 │ │ │ wang_sc │ 3 │ │ │ xin_sc │ 4 │ └────────────────────┴────────────────┴─────────────────────┘
labels State Registry ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['_scvi_labels'] │ 0 │ 0 │ └───────────────────────────┴────────────┴─────────────────────┘
INFO: GPU available: True (cuda), used: True
INFO:lightning.pytorch.utilities.rank_zero:GPU available: True (cuda), used: True
INFO: TPU available: False, using: 0 TPU cores
INFO:lightning.pytorch.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO: IPU available: False, using: 0 IPUs
INFO:lightning.pytorch.utilities.rank_zero:IPU available: False, using: 0 IPUs
INFO: HPU available: False, using: 0 HPUs
INFO:lightning.pytorch.utilities.rank_zero:HPU available: False, using: 0 HPUs
INFO: LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
INFO:lightning.pytorch.accelerators.cuda:LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 400/400: 100%|██████████| 400/400 [11:06<00:00, 1.67s/it, v_num=1, train_loss_step=1.18e+4, train_loss_epoch=1.19e+4]
INFO: `Trainer.fit` stopped: `max_epochs=400` reached.
INFO:lightning.pytorch.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=400` reached.
Epoch 400/400: 100%|██████████| 400/400 [11:07<00:00, 1.67s/it, v_num=1, train_loss_step=1.18e+4, train_loss_epoch=1.19e+4]

sc.pp.neighbors(adata_scvi, use_rep='X_scVI', key_added = "scvi_nn")
true = adata.obs["label"]
best = (0,0,0,0)
for res in np.arange(0.1, 3.01, 0.1):
sc.tl.louvain(adata_scvi, obsp='scvi_nn_distances', resolution = res, key_added = "louvain_scvi_"+str(res))
pred = adata_scvi.obs['louvain_scvi_'+str(res)]
nmi = normalized_mutual_info_score(pred, true, average_method="arithmetic")
label_ari = adjusted_rand_score(pred, y_)
batch_ari = adjusted_rand_score(pred, b_)
if best[1] < nmi:
best = (res, nmi, batch_ari, label_ari)
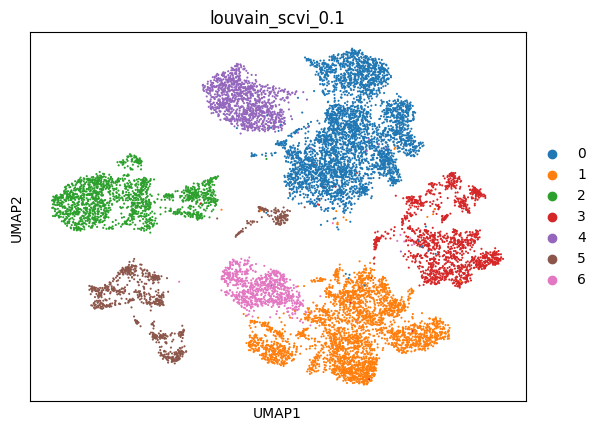
print(best)
(0.2, 0.7730430280704942, 0.008026806741679389, 0.616263135347095)
sc.tl.umap(adata_scvi)
sc.pl.umap(adata_scvi, color=['louvain_scvi_0.1'])
scVI with raw data#
X_, y_, b_, file_names = h5_data_loader(datasets, label_filter)
logging.info(f'Data loaded. {datasets}')
adata = ad.AnnData(X_, dtype=np.int32)
adata.obs["label"] = y_
adata.obs["batch"] = b_
batch_key = "batch"
label_key = "label"
adata.uns[batch_key + "_colors"] = [
"#1b9e77",
"#d95f02",
"#7570b3",
"#999999",
"#ff00ff",
] # Set custom colours for batches
sc.pp.filter_genes(adata, min_cells=2)
adata.layers["counts"] = adata.X.copy()
#sc.pp.normalize_total(adata)
#sc.pp.log1p(adata)
adata.layers["logcounts"] = adata.X.copy()
sc.tl.pca(adata)
#sc.pp.neighbors(adata)
adata_scvi = adata.copy()
scvi.model.SCVI.setup_anndata(adata_scvi, layer="counts", batch_key=batch_key)
model_scvi = scvi.model.SCVI(adata_scvi)
model_scvi.view_anndata_setup()
max_epochs_scvi = np.min([round((20000 / adata.n_obs) * 400), 400])
model_scvi.train()
adata_scvi.obsm["X_scVI"] = model_scvi.get_latent_representation()
sc.pp.neighbors(adata_scvi, use_rep="X_scVI")
sc.tl.umap(adata_scvi)
adata_scvi
sc.pl.umap(adata_scvi, color=[label_key, batch_key], wspace=1)
Anndata setup with scvi-tools version 1.0.3.
Setup via `SCVI.setup_anndata` with arguments:
{ │ 'layer': 'counts', │ 'batch_key': 'batch', │ 'labels_key': None, │ 'size_factor_key': None, │ 'categorical_covariate_keys': None, │ 'continuous_covariate_keys': None }
Summary Statistics ┏━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━┓ ┃ Summary Stat Key ┃ Value ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━┩ │ n_batch │ 5 │ │ n_cells │ 12927 │ │ n_extra_categorical_covs │ 0 │ │ n_extra_continuous_covs │ 0 │ │ n_labels │ 1 │ │ n_vars │ 15590 │ └──────────────────────────┴───────┘
Data Registry ┏━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Registry Key ┃ scvi-tools Location ┃ ┡━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ X │ adata.layers['counts'] │ │ batch │ adata.obs['_scvi_batch'] │ │ labels │ adata.obs['_scvi_labels'] │ └──────────────┴───────────────────────────┘
batch State Registry ┏━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['batch'] │ baron_sc │ 0 │ │ │ muraro_sc │ 1 │ │ │ segerstolpe_sc │ 2 │ │ │ wang_sc │ 3 │ │ │ xin_sc │ 4 │ └────────────────────┴────────────────┴─────────────────────┘
labels State Registry ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['_scvi_labels'] │ 0 │ 0 │ └───────────────────────────┴────────────┴─────────────────────┘
INFO: GPU available: True (cuda), used: True
INFO:lightning.pytorch.utilities.rank_zero:GPU available: True (cuda), used: True
INFO: TPU available: False, using: 0 TPU cores
INFO:lightning.pytorch.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO: IPU available: False, using: 0 IPUs
INFO:lightning.pytorch.utilities.rank_zero:IPU available: False, using: 0 IPUs
INFO: HPU available: False, using: 0 HPUs
INFO:lightning.pytorch.utilities.rank_zero:HPU available: False, using: 0 HPUs
INFO: LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
INFO:lightning.pytorch.accelerators.cuda:LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 400/400: 100%|██████████| 400/400 [10:44<00:00, 1.59s/it, v_num=1, train_loss_step=1.1e+4, train_loss_epoch=1.18e+4]
INFO: `Trainer.fit` stopped: `max_epochs=400` reached.
INFO:lightning.pytorch.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=400` reached.
Epoch 400/400: 100%|██████████| 400/400 [10:44<00:00, 1.61s/it, v_num=1, train_loss_step=1.1e+4, train_loss_epoch=1.18e+4]

sc.pp.neighbors(adata_scvi, use_rep='X_scVI', key_added = "scvi_nn")
true = adata.obs["label"]
best = (0,0,0,0)
for res in np.arange(0.1, 3.01, 0.1):
sc.tl.louvain(adata_scvi, obsp='scvi_nn_distances', resolution = res, key_added = "louvain_scvi_"+str(res))
pred = adata_scvi.obs['louvain_scvi_'+str(res)]
nmi = normalized_mutual_info_score(pred, true, average_method="arithmetic")
label_ari = adjusted_rand_score(pred, y_)
batch_ari = adjusted_rand_score(pred, b_)
if best[1] < nmi:
best = (res, nmi, batch_ari, label_ari)
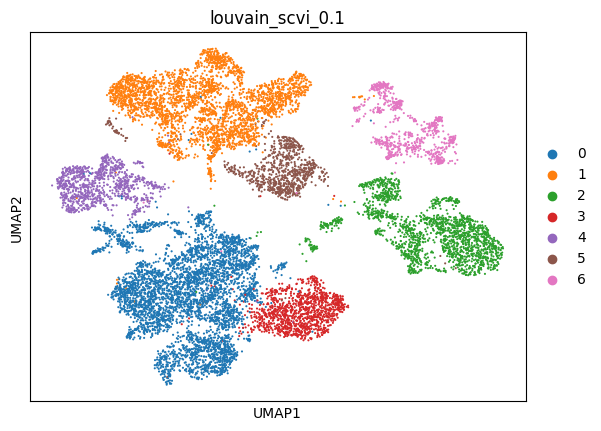
print(best)
(0.1, 0.7470742452907168, -0.03283466027592697, 0.653151410586291)
sc.tl.umap(adata_scvi)
sc.pl.umap(adata_scvi, color=['louvain_scvi_0.1'])
scANVI with Total->Log preprocessing#
|model_scanvi = scvi.model.SCANVI.from_scvi_model(
model_scvi, labels_key=label_key, unlabeled_category="unlabelled"
)
print(model_scanvi)
model_scanvi.view_anndata_setup()
ScanVI Model with the following params: unlabeled_category: unlabelled, n_hidden: 128, n_latent: 10, n_layers: 1, dropout_rate: 0.1, dispersion: gene, gene_likelihood: zinb Training status: Not Trained Model's adata is minified?: False
Anndata setup with scvi-tools version 1.0.3.
Setup via `SCANVI.setup_anndata` with arguments:
{ │ 'labels_key': 'label', │ 'unlabeled_category': 'unlabelled', │ 'layer': 'counts', │ 'batch_key': 'batch', │ 'size_factor_key': None, │ 'categorical_covariate_keys': None, │ 'continuous_covariate_keys': None }
Summary Statistics ┏━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━┓ ┃ Summary Stat Key ┃ Value ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━┩ │ n_batch │ 5 │ │ n_cells │ 12927 │ │ n_extra_categorical_covs │ 0 │ │ n_extra_continuous_covs │ 0 │ │ n_labels │ 14 │ │ n_vars │ 15590 │ └──────────────────────────┴───────┘
Data Registry ┏━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Registry Key ┃ scvi-tools Location ┃ ┡━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ X │ adata.layers['counts'] │ │ batch │ adata.obs['_scvi_batch'] │ │ labels │ adata.obs['_scvi_labels'] │ └──────────────┴───────────────────────────┘
batch State Registry ┏━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['batch'] │ baron_sc │ 0 │ │ │ muraro_sc │ 1 │ │ │ segerstolpe_sc │ 2 │ │ │ wang_sc │ 3 │ │ │ xin_sc │ 4 │ └────────────────────┴────────────────┴─────────────────────┘
labels State Registry ┏━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['label'] │ acinar │ 0 │ │ │ activated │ 1 │ │ │ alpha │ 2 │ │ │ beta │ 3 │ │ │ delta │ 4 │ │ │ duct │ 5 │ │ │ endothelial │ 6 │ │ │ epsilon │ 7 │ │ │ gamma │ 8 │ │ │ macrophage │ 9 │ │ │ mast │ 10 │ │ │ quiescent │ 11 │ │ │ schwann │ 12 │ │ │ unlabelled │ 13 │ └────────────────────┴─────────────┴─────────────────────┘
max_epochs_scanvi = int(np.min([10, np.max([2, round(max_epochs_scvi / 3.0)])]))
model_scanvi.train(max_epochs=max_epochs_scanvi)
INFO Training for 10 epochs.
INFO: GPU available: True (cuda), used: True
INFO:lightning.pytorch.utilities.rank_zero:GPU available: True (cuda), used: True
INFO: TPU available: False, using: 0 TPU cores
INFO:lightning.pytorch.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO: IPU available: False, using: 0 IPUs
INFO:lightning.pytorch.utilities.rank_zero:IPU available: False, using: 0 IPUs
INFO: HPU available: False, using: 0 HPUs
INFO:lightning.pytorch.utilities.rank_zero:HPU available: False, using: 0 HPUs
INFO: LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
INFO:lightning.pytorch.accelerators.cuda:LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 10/10: 100%|██████████| 10/10 [00:50<00:00, 4.62s/it, v_num=1, train_loss_step=9.78e+3, train_loss_epoch=1.2e+4]
INFO: `Trainer.fit` stopped: `max_epochs=10` reached.
INFO:lightning.pytorch.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=10` reached.
Epoch 10/10: 100%|██████████| 10/10 [00:50<00:00, 5.05s/it, v_num=1, train_loss_step=9.78e+3, train_loss_epoch=1.2e+4]
adata_scanvi = adata_scvi.copy()
adata_scanvi.obsm["X_scANVI"] = model_scanvi.get_latent_representation()
sc.pp.neighbors(adata_scanvi, use_rep="X_scANVI")
sc.tl.umap(adata_scanvi)
sc.pl.umap(adata_scanvi, color=[label_key, batch_key], wspace=1)

sc.pp.neighbors(adata_scanvi, use_rep='X_scANVI', key_added = "scanvi_nn")
true = adata.obs["label"]
best = (0,0,0,0)
for res in np.arange(0.1, 3.01, 0.1):
sc.tl.louvain(adata_scanvi, obsp='scanvi_nn_distances', resolution = res, key_added = "louvain_scanvi_"+str(res))
pred = adata_scanvi.obs['louvain_scanvi_'+str(res)]
nmi = normalized_mutual_info_score(pred, true, average_method="arithmetic")
label_ari = adjusted_rand_score(pred, y_)
batch_ari = adjusted_rand_score(pred, b_)
if best[1] < nmi:
best = (res, nmi, batch_ari, label_ari)
print(best)
(0.1, 0.8733531457859234, -0.019799213315938195, 0.9367992377003943)
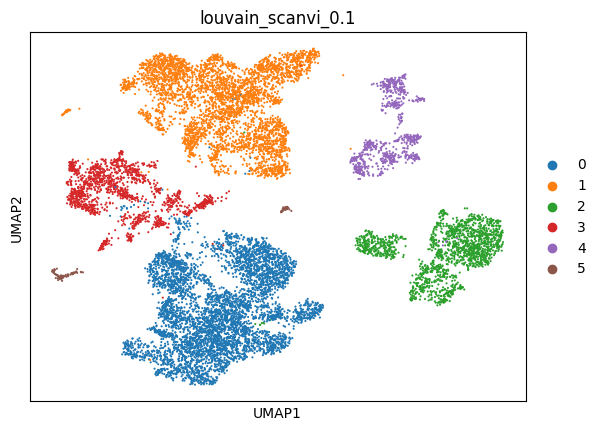
sc.tl.umap(adata_scanvi)
sc.pl.umap(adata_scanvi, color=['louvain_scanvi_0.1'])
BBKNN#
!pip install --quiet bbknn
import bbknn
adata_bbknn = adata.copy()
adata_bbknn.X = adata_bbknn.layers["logcounts"].copy()
sc.pp.pca(adata_bbknn)
bbknn.bbknn(
adata_bbknn, batch_key=batch_key, neighbors_within_batch=3
)
adata_bbknn
AnnData object with n_obs × n_vars = 12927 × 15590
obs: 'label', 'batch'
var: 'n_cells'
uns: 'batch_colors', 'log1p', 'pca', 'neighbors'
obsm: 'X_pca'
varm: 'PCs'
layers: 'counts', 'logcounts'
obsp: 'distances', 'connectivities'
sc.tl.umap(adata_bbknn)
sc.pl.umap(adata_bbknn, color=[label_key, batch_key], wspace=1)